7N50
 
 | |
7N52
 
 | |
7N51
 
 | |
6WKN
 
 | | PL-bound rat TRPV2 in nanodiscs | | Descriptor: | Transient receptor potential cation channel subfamily V member 2, piperlongumine | | Authors: | Pumroy, R.P, Moiseenkova-Bell, V.Y. | | Deposit date: | 2020-04-16 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Allosteric Antagonist Modulation of TRPV2 by Piperlongumine Impairs Glioblastoma Progression.
Acs Cent.Sci., 7, 2021
|
|
8EKD
 
 | |
7QUE
 
 | | The STK17A (DRAK1) Kinase Domain Bound to CKJB68 | | Descriptor: | Serine/threonine-protein kinase 17A, ~{N}-(phenylmethyl)-7,10-dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.1^{2,6}.0^{17,20}]docosa-1(20),2(22),3,5,14(21),15,18-heptaene-5-carboxamide | | Authors: | Mathea, S, Preuss, F, Chatterjee, D, Dederer, V, Kurz, C.G, Amrhein, J.A, Hanke, T, Knapp, S. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Illuminating the Dark: Highly Selective Inhibition of Serine/Threonine Kinase 17A with Pyrazolo[1,5- a ]pyrimidine-Based Macrocycles.
J.Med.Chem., 65, 2022
|
|
7QUF
 
 | | The STK17A (DRAK1) Kinase Domain Bound to CK156 | | Descriptor: | Serine/threonine-protein kinase 17A, ~{N}-~{tert}-butyl-7,10-dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.1^{2,6}.0^{17,20}]docosa-1(20),2(22),3,5,14(21),15,18-heptaene-5-carboxamide | | Authors: | Mathea, S, Preuss, F, Chatterjee, D, Dederer, V, Kurz, C, Hanke, T, Knapp, S. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Illuminating the Dark: Highly Selective Inhibition of Serine/Threonine Kinase 17A with Pyrazolo[1,5- a ]pyrimidine-Based Macrocycles.
J.Med.Chem., 65, 2022
|
|
7QUT
 
 | | serial synchrotron crystallographic structure of Drosophila Melanogaster (6-4) photolyase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, RE11660p | | Authors: | Cellini, A, Weixiao, Y.W, Kumar, M.S, Westenhoff, S. | | Deposit date: | 2022-01-18 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis of the radical pair state in photolyases and cryptochromes.
Chem.Commun.(Camb.), 58, 2022
|
|
7AT6
 
 | | Structure of thaumatin collected by femtosecond serial crystallography on a COC membrane | | Descriptor: | L(+)-TARTARIC ACID, R-1,2-PROPANEDIOL, SODIUM ION, ... | | Authors: | Martiel, I, Marsh, M, Vera, L, Huang, C.Y, Olieric, V, Leonarski, P, Nass, K, Padeste, C, Karpik, A, Wang, M, Pedrini, B. | | Deposit date: | 2020-10-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Commissioning results from the SwissMX instrument for fixed target macromolecular crystallography at SwissFEL
To Be Published
|
|
7PUE
 
 | | Human serum and glucocorticoid-regulated kinase 1 in complex with pyrazolopyridine inhibitor 3a | | Descriptor: | 6-[4-[[2,3-bis(chloranyl)phenyl]sulfonylamino]phenyl]-~{N}-[(3~{R})-pyrrolidin-3-yl]-2~{H}-pyrazolo[3,4-b]pyridine-4-carboxamide, GLYCEROL, Serine/threonine-protein kinase Sgk1 | | Authors: | Dreyer, M.K, Halland, N, Nazare, M. | | Deposit date: | 2021-09-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Rational Design of Highly Potent, Selective, and Bioavailable SGK1 Protein Kinase Inhibitors for the Treatment of Osteoarthritis.
J.Med.Chem., 65, 2022
|
|
5O5K
 
 | | X-ray structure of a bacterial adenylyl cyclase soluble domain | | Descriptor: | 3'-O-(N-METHYLANTHRANILOYL)-GUANOSINE-5'-TRIPHOSPHATE, Adenylate cyclase, MANGANESE (II) ION, ... | | Authors: | Vercellino, I, Korkhov, V.M. | | Deposit date: | 2017-06-02 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Role of the nucleotidyl cyclase helical domain in catalytically active dimer formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8SG1
 
 | | Cryo-EM structure of CMKLR1 signaling complex | | Descriptor: | CHOLESTEROL, Chemerin 9, Chemerin-like receptor 1, ... | | Authors: | Zhang, X, Zhang, C. | | Deposit date: | 2023-04-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis of G protein-Coupled receptor CMKLR1 activation and signaling induced by a chemerin-derived agonist.
Plos Biol., 21, 2023
|
|
8B4I
 
 | | Cryo-EM structure of the Neurospora crassa TOM core complex at 3.3 angstrom | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, DIUNDECYL PHOSPHATIDYL CHOLINE, Mitochondrial import receptor subunit Tom22, ... | | Authors: | Ornelas, P, Kuehlbrandt, W. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Two conformations of the Tom20 preprotein receptor in the TOM holo complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7PBC
 
 | | Crystal structure of engineered TCR (796) complexed to HLA-A*02:01 presenting MAGE-A10 9-mer peptide | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GLYCEROL, ... | | Authors: | Simister, P.C, Border, E.C, Vieira, J.F, Pumphrey, N.J. | | Deposit date: | 2021-08-02 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural insights into engineering a T-cell receptor targeting MAGE-A10 with higher affinity and specificity for cancer immunotherapy.
J Immunother Cancer, 10, 2022
|
|
4DBM
 
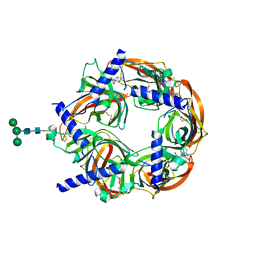 | | Aplysia californica-AChBP in complex with triazole 18 | | Descriptor: | (3-exo)-8,8-dimethyl-3-(4-{[(1-methyl-2-oxo-1,2-dihydroquinolin-4-yl)oxy]methyl}-1H-1,2,3-triazol-1-yl)-8-azoniabicyclo[3.2.1]octane, 2-acetamido-2-deoxy-beta-D-glucopyranose, Soluble acetylcholine receptor, ... | | Authors: | Nemecz, A, Yamauchi, J.G, Kim, C. | | Deposit date: | 2012-01-16 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Generation of candidate ligands for nicotinic acetylcholine receptors via in situ click chemistry with a soluble acetylcholine binding protein template.
J.Am.Chem.Soc., 134, 2012
|
|
8P1B
 
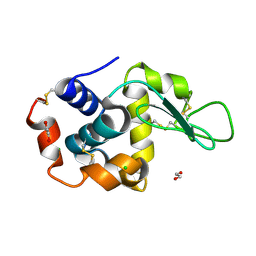 | | Lysozyme structure solved from serial crystallography data collected at 2 kHz with JUNGFRAU detector at MAXIV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Nan, J, Leonarski, F, Furrer, A, Dworkowski, F. | | Deposit date: | 2023-05-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kilohertz serial crystallography with the JUNGFRAU detector at a fourth-generation synchrotron source.
Iucrj, 10, 2023
|
|
8P1C
 
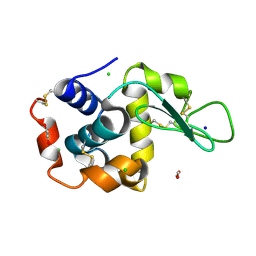 | | Lysozyme structure solved from serial crystallography data collected at 1 kHz with JUNGFRAU detector at MAXIV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Nan, J, Leonarski, F, Furrer, A, Dworkowski, F. | | Deposit date: | 2023-05-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Kilohertz serial crystallography with the JUNGFRAU detector at a fourth-generation synchrotron source.
Iucrj, 10, 2023
|
|
8P1A
 
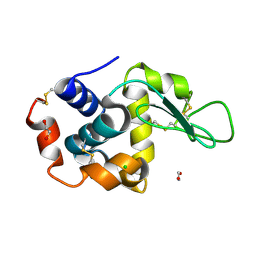 | | Lysozyme structure solved from serial crystallography data collected at 2 kHz for 5 seconds with JUNGFRAU detector at MAXIV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C | | Authors: | Nan, J, Leonarski, F, Furrer, A, Dworkowski, F. | | Deposit date: | 2023-05-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Kilohertz serial crystallography with the JUNGFRAU detector at a fourth-generation synchrotron source.
Iucrj, 10, 2023
|
|
8P1D
 
 | | Lysozyme structure solved from serial crystallography data collected at 100 Hz with JUNGFRAU detector at MAXIV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Nan, J, Leonarski, F, Furrer, A, Dworkowski, F. | | Deposit date: | 2023-05-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Kilohertz serial crystallography with the JUNGFRAU detector at a fourth-generation synchrotron source.
Iucrj, 10, 2023
|
|
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | Descriptor: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
5D59
 
 | | In meso X-ray crystallography structure of the PepTSt-Ala-Phe complex at 100 K | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ALANINE, ... | | Authors: | Huang, C.-Y, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D5C
 
 | | In meso in situ serial X-ray crystallography structure of lysozyme at 100 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETIC ACID, BROMIDE ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D52
 
 | | In meso in situ serial X-ray crystallography structure of insulin at room temperature | | Descriptor: | Insulin A chain, Insulin B chain, PHOSPHATE ION | | Authors: | Huang, C.-Y, Olieric, V, Warshamanage, R, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D54
 
 | | In meso X-ray crystallography structure of insulin at 100 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Insulin A chain, Insulin B chain, ... | | Authors: | Huang, C.-Y, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D57
 
 | | In meso X-ray crystallography structure of diacylglycerol kinase, DgkA, at 100 K | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Diacylglycerol kinase | | Authors: | Huang, C.-Y, Howe, N, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
