8I68
 
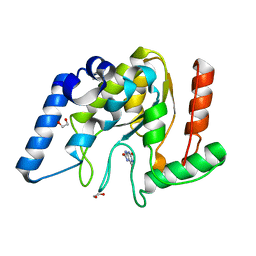 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Uric acid, Form III | | Descriptor: | 1,2-ETHANEDIOL, URIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
6Q9Z
 
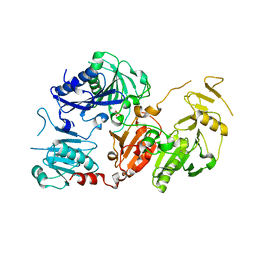 | | Crystal structure of the pathological G167R variant of calcium-free human gelsolin, | | Descriptor: | GLYCEROL, Gelsolin, SULFATE ION | | Authors: | Boni, F, Scalone, E, Milani, M, Eloise, M, de Rosa, M. | | Deposit date: | 2018-12-18 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
6QBF
 
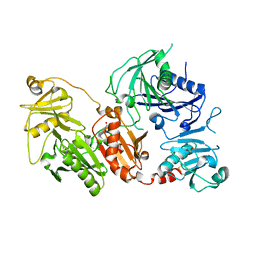 | | Crystal structure of the pathological D187N variant of calcium-free human gelsolin. | | Descriptor: | GLYCEROL, Gelsolin, SODIUM ION, ... | | Authors: | Scalone, E, Boni, F, Milani, M, Eloise, M, de Rosa, M. | | Deposit date: | 2018-12-21 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.499 Å) | | Cite: | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
6Q9R
 
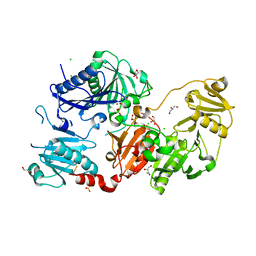 | | Crystal structure of the pathological N184K variant of calcium-free human gelsolin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Scalone, E, Boni, F, Milani, M, Eloise, M, de Rosa, M. | | Deposit date: | 2018-12-18 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
5MTL
 
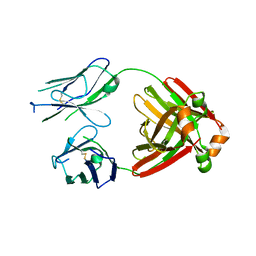 | | Crystal structure of an amyloidogenic light chain | | Descriptor: | light chain dimer,IGL@ protein,IGL@ protein | | Authors: | Oberti, L, Rognoni, P, Russo, R, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-10 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
7VRQ
 
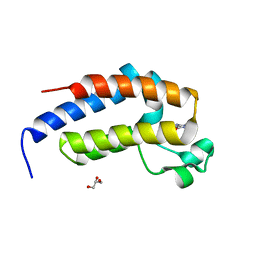 | | crystal structure of BRD2-BD2 in complex with purine derivative | | Descriptor: | Bromodomain-containing protein 2, GLYCEROL, THEOBROMINE | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-23 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VS0
 
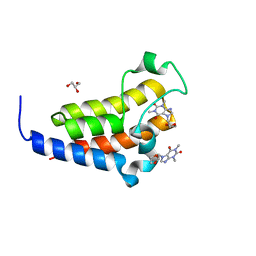 | | crystal structure of BRD2-BD2 in complex with purine derivative | | Descriptor: | Bromodomain-containing protein 2, Doxofylline, GLYCEROL | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VRM
 
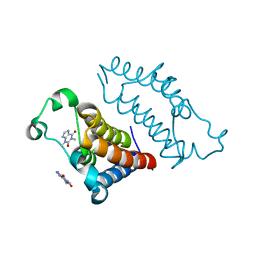 | | crystal structure of BRD2-BD2 in complex with purine derivative | | Descriptor: | Bromodomain-containing protein 2, THEOPHYLLINE | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-23 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VRO
 
 | | crystal structure of BRD2-BD1 in complex with purine derivative | | Descriptor: | Bromodomain-containing protein 2, SULFATE ION, THEOBROMINE | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-23 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VRK
 
 | | crystal structure of BRD2-BD1 in complex with purine derivative | | Descriptor: | Bromodomain-containing protein 2, SULFATE ION, THEOPHYLLINE | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-23 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VRH
 
 | | crystal structure of BRD2-BD1 in complex with guanosine analog | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 9-HYROXYETHOXYMETHYLGUANINE, Bromodomain-containing protein 2, ... | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-22 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VS1
 
 | | crystal structure of BRD2-BD2 in complex with purine derivative | | Descriptor: | 3-methyl-7-propyl-purine-2,6-dione, Bromodomain-containing protein 2, GLYCEROL | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VSF
 
 | | crystal structure of BRD2-BD1 in complex with purine derivative | | Descriptor: | 3-methyl-7-propyl-purine-2,6-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bromodomain-containing protein 2, ... | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-26 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VRI
 
 | | crystal structure of BRD2-BD2 in complex with guanosine analog | | Descriptor: | 9-HYROXYETHOXYMETHYLGUANINE, Bromodomain-containing protein 2 | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-23 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VRZ
 
 | | crystal structure of BRD2-BD1 in complex with purine derivative | | Descriptor: | Bromodomain-containing protein 2, Doxofylline, SULFATE ION | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5MUD
 
 | | Crystal structure of an amyloidogenic light chain dimer H6 | | Descriptor: | light chain dimer,IGL@ protein | | Authors: | Oberti, L, Bacarizo, J, Maritan, M, Rognoni, P, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
6I8C
 
 | | Crystal structure of the murine beta-2-microglobulin. | | Descriptor: | Beta-2-microglobulin, TRIETHYLENE GLYCOL | | Authors: | Achour, A, Sandalova, T, Ricagno, S, Sun, R. | | Deposit date: | 2018-11-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Biochemical and biophysical comparison of human and mouse beta-2 microglobulin reveals the molecular determinants of low amyloid propensity.
Febs J., 287, 2020
|
|
5O2Z
 
 | | Domain swap dimer of the G167R variant of gelsolin second domain | | Descriptor: | ACETATE ION, CALCIUM ION, CITRATE ANION, ... | | Authors: | Boni, F, Milani, M, Mastrangelo, E, de Rosa, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Gelsolin pathogenic Gly167Arg mutation promotes domain-swap dimerization of the protein.
Hum. Mol. Genet., 27, 2018
|
|
5M76
 
 | | Crystal structure of cardiotoxic Bence-Jones light chain dimer H10 | | Descriptor: | BROMIDE ION, light chain dimer | | Authors: | Oberti, L, Rognoni, P, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2016-10-26 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
6FXN
 
 | | Crystal structure of human BAFF in complex with Fab fragment of anti-BAFF antibody belimumab | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, belimumab heavy chain, belimumab light chain | | Authors: | Lammens, A, Maskos, K, Willen, L, Jiang, X, Schneider, P. | | Deposit date: | 2018-03-09 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A loop region of BAFF controls B cell survival and regulates recognition by different inhibitors.
Nat Commun, 9, 2018
|
|
6H1F
 
 | | Structure of the nanobody-stabilized gelsolin D187N variant (second domain) | | Descriptor: | Gelsolin, THIOCYANATE ION, gelsolin nanobody, ... | | Authors: | Hassan, A, Milani, M, Mastrangelo, E, de Rosa, M. | | Deposit date: | 2018-07-11 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nanobody interaction unveils structure, dynamics and proteotoxicity of the Finnish-type amyloidogenic gelsolin variant.
Biochim Biophys Acta Mol Basis Dis, 1865, 2019
|
|
5MUH
 
 | | Crystal structure of an amyloidogenic light chain dimer H7 | | Descriptor: | light chain dimer | | Authors: | Oberti, L, Rognoni, P, Russo, R, Maritan, M, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
5M6A
 
 | | Crystal structure of cardiotoxic Bence-Jones light chain dimer H9 | | Descriptor: | Bence-Jones light chain, GLYCEROL, PHOSPHATE ION | | Authors: | Oberti, L, Rognoni, P, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2016-10-24 | | Release date: | 2017-11-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
5M6I
 
 | | Crystal structure of non-cardiotoxic Bence-Jones light chain dimer M8 | | Descriptor: | SODIUM ION, light chain dimer | | Authors: | Oberti, L, Rognoni, P, Russo, R, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2016-10-25 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
7ENZ
 
 | | Crystal structure of Phenanthredinone moiety in complex with the second bromodomain of BRD2 (BRD2-BD2). | | Descriptor: | Bromodomain-containing protein 2, TRIETHYLENE GLYCOL, phenanthridin-6(5H)-one | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural investigation of a pyrano-1,3-oxazine derivative and the phenanthridinone core moiety against BRD2 bromodomains.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
