6X9J
 
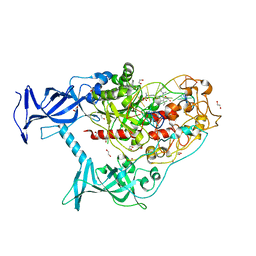 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3830052 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*CP*(5CM)P*GP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*G)-R(P*(PYO))-D(P*GP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Pathuri, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-06-02 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia.
Nat Cancer, 2, 2021
|
|
6X9I
 
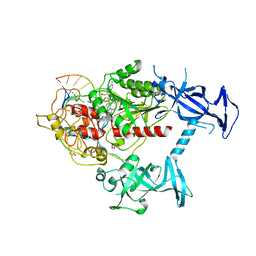 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*CP*(5CM)P*GP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*G)-R(P*(PYO))-D(P*GP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Pathuri, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-06-02 | | Release date: | 2021-07-07 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia.
Nat Cancer, 2, 2021
|
|
8FS1
 
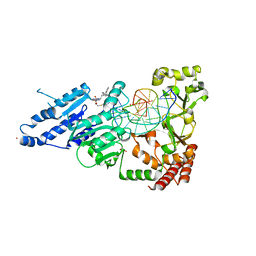 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor 11a (YD905) | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-{2-[N'-(cyclohexylmethyl)carbamimidamido]ethyl}-N-(3-phenylpropyl)-5'-thioadenosine, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-01-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Comparative Study of Adenosine Analogs as Inhibitors of Protein Arginine Methyltransferases and a Clostridioides difficile- Specific DNA Adenine Methyltransferase.
Acs Chem.Biol., 18, 2023
|
|
8FS2
 
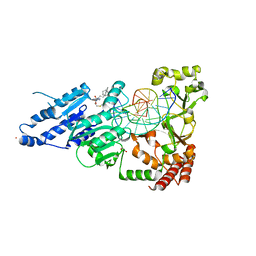 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor 11b (YD907) | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-{3-[N'-(cyclohexylmethyl)carbamimidamido]propyl}-N-(3-phenylpropyl)-5'-thioadenosine, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-01-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Comparative Study of Adenosine Analogs as Inhibitors of Protein Arginine Methyltransferases and a Clostridioides difficile- Specific DNA Adenine Methyltransferase.
Acs Chem.Biol., 18, 2023
|
|
1ML9
 
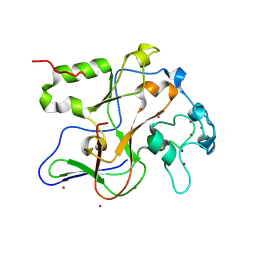 | | Structure of the Neurospora SET domain protein DIM-5, a histone lysine methyltransferase | | Descriptor: | Histone H3 methyltransferase DIM-5, UNKNOWN LIGAND, ZINC ION | | Authors: | Zhang, X, Tamaru, H, Khan, S.I, Horton, J.R, Keefe, L.J, Selker, E.U, Cheng, X. | | Deposit date: | 2002-08-30 | | Release date: | 2002-10-23 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the Neurospora SET domain protein DIM-5,
a histone H3 lysine methyltransferase
Cell(Cambridge,Mass.), 111, 2002
|
|
1MOZ
 
 | | ADP-ribosylation factor-like 1 (ARL1) from Saccharomyces cerevisiae | | Descriptor: | ADP-ribosylation factor-like protein 1, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Amor, J.C, Horton, J.R, Zhu, X, Wang, Y, Sullards, C, Ringe, D, Cheng, X, Kahn, R.A. | | Deposit date: | 2002-09-10 | | Release date: | 2002-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structures of Yeast ARF2 and ARL1:
DISTINCT ROLES FOR THE N TERMINUS IN THE STRUCTURE
AND FUNCTION OF ARF FAMILY GTPases
J.Biol.Chem., 276, 2001
|
|
1MR3
 
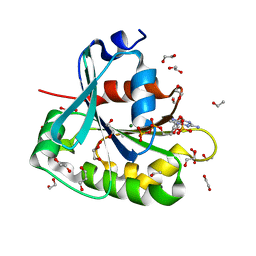 | | Saccharomyces cerevisiae ADP-ribosylation Factor 2 (ScArf2) complexed with GDP-3'P at 1.6A resolution | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ADP-ribosylation factor 2, ... | | Authors: | Amor, J.-C, Horton, J.R, Zhu, X, Wang, Y, Sullards, C, Ringe, D, Cheng, X, Kahn, R.A. | | Deposit date: | 2002-09-17 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of yeast ARF2 and ARL1: distinct roles for the N terminus in the structure and function of ARF family GTPases.
J.Biol.Chem., 276, 2001
|
|
3MO2
 
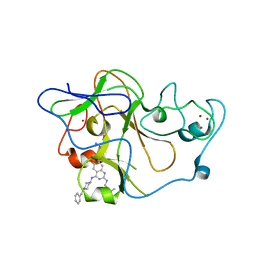 | | human G9a-like (GLP, also known as EHMT1) in complex with inhibitor E67 | | Descriptor: | 7-[(5-aminopentyl)oxy]-N~4~-(1-benzylpiperidin-4-yl)-N~2~-[3-(dimethylamino)propyl]-6-methoxyquinazoline-2,4-diamine, Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, ... | | Authors: | Chang, Y, Horton, J.R, Cheng, X. | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases.
J.Mol.Biol., 400, 2010
|
|
3MO5
 
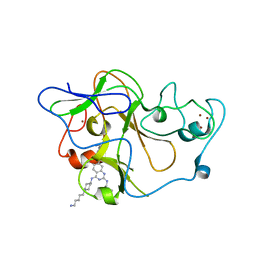 | | Human G9a-like (GLP, also known as EHMT1) in complex with inhibitor E72 | | Descriptor: | 7-[(5-aminopentyl)oxy]-N~4~-[1-(5-aminopentyl)piperidin-4-yl]-N~2~-[3-(dimethylamino)propyl]-6-methoxyquinazoline-2,4-diamine, Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, ... | | Authors: | Chang, Y, Horton, J.R, Cheng, X. | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases.
J.Mol.Biol., 400, 2010
|
|
3MO0
 
 | | Human G9a-like (GLP, also known as EHMT1) in complex with inhibitor E11 | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, ... | | Authors: | Chang, Y, Horton, J.R, Cheng, X. | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases.
J.Mol.Biol., 400, 2010
|
|
1YNM
 
 | | Crystal structure of restriction endonuclease HinP1I | | Descriptor: | R.HinP1I restriction endonuclease | | Authors: | Yang, Z, Horton, J.R, Maunus, R, Wilson, G.G, Roberts, R.J, Cheng, X. | | Deposit date: | 2005-01-24 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of HinP1I endonuclease reveals a striking similarity to the monomeric restriction enzyme MspI
Nucleic Acids Res., 33, 2005
|
|
3QXY
 
 | | Human SETD6 in complex with RelA Lys310 | | Descriptor: | 1,2-ETHANEDIOL, N-lysine methyltransferase SETD6, S-ADENOSYLMETHIONINE, ... | | Authors: | Chang, Y, Levy, D, Horton, J.R, Peng, J, Zhang, X, Gozani, O, Cheng, X. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural basis of SETD6-mediated regulation of the NF-kB network via methyl-lysine signaling.
Nucleic Acids Res., 39, 2011
|
|
3OS5
 
 | | SET7/9-Dnmt1 K142me1 complex | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Dnmt1, ... | | Authors: | Esteve, P.-O, Chang, Y, Samaranayake, M, Upadhyay, A.K, Horton, J.R, Feehery, G.R, Cheng, X, Pradhan, S. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3RC0
 
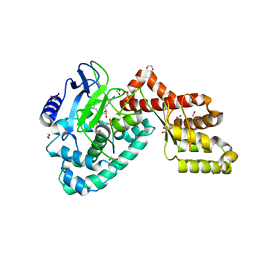 | | Human SETD6 in complex with RelA Lys310 peptide | | Descriptor: | 1,2-ETHANEDIOL, N-lysine methyltransferase SETD6, S-ADENOSYLMETHIONINE, ... | | Authors: | Chang, Y, Levy, D, Horton, J.R, Peng, J, Zhang, X, Gozani, O, Cheng, X. | | Deposit date: | 2011-03-30 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis of SETD6-mediated regulation of the NF-kB network via methyl-lysine signaling.
Nucleic Acids Res., 39, 2011
|
|
3QO2
 
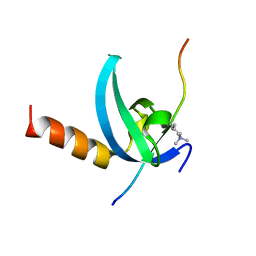 | | Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9 | | Descriptor: | 1,2-ETHANEDIOL, Histone H3 peptide, M-phase phosphoprotein 8 | | Authors: | Chang, Y, Horton, J.R, Bedford, M.T, Zhang, X, Cheng, X. | | Deposit date: | 2011-02-09 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9: potential effect of phosphorylation on methyl-lysine binding.
J.Mol.Biol., 408, 2011
|
|
7MHT
 
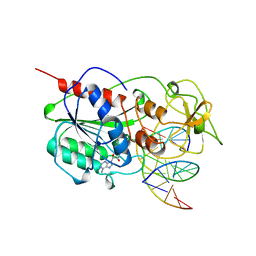 | | CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI/DNA COMPLEX | | Descriptor: | 5'-D(P*CP*CP*AP*TP*GP*AP*GP*CP*TP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3', CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI, ... | | Authors: | O'Gara, M, Horton, J.R, Roberts, R.J, Cheng, X. | | Deposit date: | 1998-08-05 | | Release date: | 1998-11-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base.
Nat.Struct.Biol., 5, 1998
|
|
1Q0T
 
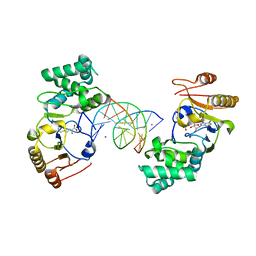 | | Ternary Structure of T4DAM with AdoHcy and DNA | | Descriptor: | 5'-D(*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*T)-3', DNA adenine methylase, IODIDE ION, ... | | Authors: | Yang, Z, Horton, J.R, Zhou, L, Zhang, X.J, Dong, A, Zhang, X, Schlagman, S.L, Kossykh, V, Hattman, S, Cheng, X. | | Deposit date: | 2003-07-17 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the bacteriophage T4 DNA adenine methyltransferase
Nat.Struct.Biol., 10, 2003
|
|
2G45
 
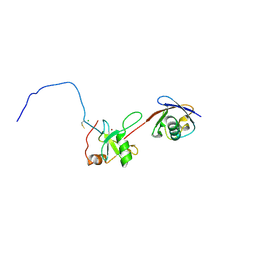 | | Co-crystal structure of znf ubp domain from the deubiquitinating enzyme isopeptidase T (isot) in complex with ubiquitin | | Descriptor: | CHLORIDE ION, Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 5, ... | | Authors: | Reyes-Turcu, F.E, Horton, J.R, Mullally, J.E, Heroux, A, Cheng, X, Wilkinson, K.D. | | Deposit date: | 2006-02-21 | | Release date: | 2006-04-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Ubiquitin Binding Domain ZnF UBP Recognizes the C-Terminal Diglycine Motif of Unanchored Ubiquitin.
Cell(Cambridge,Mass.), 124, 2006
|
|
1Q0S
 
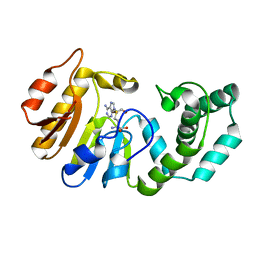 | | Binary Structure of T4DAM with AdoHcy | | Descriptor: | DNA adenine methylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yang, Z, Horton, J.R, Zhou, L, Zhang, X.J, Dong, A, Zhang, X, Schlagman, S.L, Kossykh, V, Hattman, S, Cheng, X. | | Deposit date: | 2003-07-17 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the bacteriophage T4 DNA adenine methyltransferase
Nat.Struct.Biol., 10, 2003
|
|
2G43
 
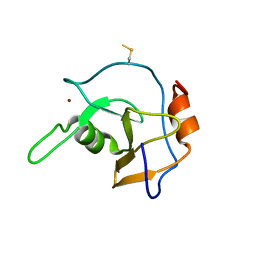 | | Structure of the ZNF UBP domain from deubiquitinating enzyme isopeptidase T (IsoT) | | Descriptor: | UNKNOWN ATOM OR ION, Ubiquitin carboxyl-terminal hydrolase 5, ZINC ION | | Authors: | Reyes-Turcu, F.E, Horton, J.R, Mullally, J.E, Heroux, A, Cheng, X, Wilkinson, K.D. | | Deposit date: | 2006-02-21 | | Release date: | 2006-04-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The Ubiquitin Binding Domain ZnF UBP Recognizes the C-Terminal Diglycine Motif of Unanchored Ubiquitin.
Cell(Cambridge,Mass.), 124, 2006
|
|
1PEG
 
 | | Structural basis for the product specificity of histone lysine methyltransferases | | Descriptor: | Histone H3, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Zhang, X, Yang, Z, Khan, S.I, Horton, J.R, Tamaru, H, Selker, E.U, Cheng, X. | | Deposit date: | 2003-05-21 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis for the product specificity of histone lysine methyltransferases
Mol.Cell, 12, 2003
|
|
1BM8
 
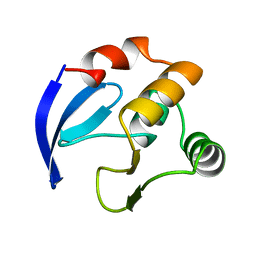 | | DNA-BINDING DOMAIN OF MBP1 | | Descriptor: | TRANSCRIPTION FACTOR MBP1 | | Authors: | Xu, R.-M, Koch, C, Liu, Y, Horton, J.R, Knapp, D, Nasmyth, K, Cheng, X. | | Deposit date: | 1998-07-29 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of the DNA-binding domain of Mbp1, a transcription factor important in cell-cycle control of DNA synthesis.
Structure, 5, 1997
|
|
8S9M
 
 | | DNA cytosine-N4 methyltransferase (residues 79-324) from the Bdelloid rotifer Adineta vaga | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
8S9O
 
 | | DNA cytosine-N4 methyltransferase (residues 61-324) from the Bdelloid rotifer Adineta vaga - P1 crystal form | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
8S9N
 
 | | DNA cytosine-N4 methyltransferase (residues 61-324) from the Bdelloid rotifer Adineta vaga - C2 crystal form | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
