6M7M
 
 | | rac-GSTSTA from degenerate octameric repeats in InaZ, residues 707-712 | | Descriptor: | L-GSTSTA from ice nucleation protein, inaZ, and its enantiomer, ... | | Authors: | Zee, C, Glynn, C, Gallagher-Jones, M, Miao, J, Santiago, C.G, Cascio, D, Gonen, T, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Homochiral and racemic MicroED structures of a peptide repeat from the ice-nucleation protein InaZ.
IUCrJ, 6, 2019
|
|
6M9J
 
 | | Racemic-GSTSTA from degenerate octameric repeats in InaZ, residues 707-712 | | Descriptor: | Ice nucleation protein | | Authors: | Zee, C, Glynn, C, Gallagher-Jones, M, Miao, J, Santiago, C.G, Cascio, D, Gonen, T, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2018-08-23 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.9 Å) | | Cite: | Homochiral and racemic MicroED structures of a peptide repeat from the ice-nucleation protein InaZ.
IUCrJ, 6, 2019
|
|
6N3C
 
 | | SegB, conformation of TDP-43 low complexity domain segment A | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-11-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of four polymorphic TDP-43 amyloid cores.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6N3B
 
 | | SegA-asym, conformation of TDP-43 low complexity domain segment A asym | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-11-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of four polymorphic TDP-43 amyloid cores.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6N3A
 
 | | SegA-long, conformation of TDP-43 low complexity domain segment A long | | Descriptor: | TAR DNA-binding protein 43, segA long small | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-11-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of four polymorphic TDP-43 amyloid cores.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6N37
 
 | | SegA-sym, conformation of TDP-43 low complexity domain segment A sym | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-11-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of four polymorphic TDP-43 amyloid cores.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NK4
 
 | |
6WPQ
 
 | | GNYNVF from hnRNPA2-low complexity domain segment, residues 286-291, D290V variant | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A2 | | Authors: | Lu, J, Cao, Q, Hughes, M.P, Sawaya, M.R, Boyer, D.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | CryoEM structure of the low-complexity domain of hnRNPA2 and its conversion to pathogenic amyloid.
Nat Commun, 11, 2020
|
|
6X1I
 
 | |
7QYD
 
 | | mosquitocidal Cry11Ba determined at pH 6.5 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Ba | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P, Sawaya, M.R, Schibrowsky, N.A, Cascio, D, Rodriguez, J.A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
6XPH
 
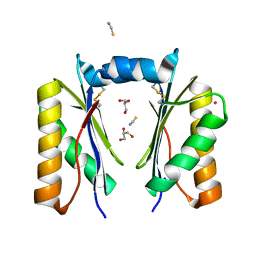 | | CutR dimer with domain swap | | Descriptor: | Ethanolamine utilization protein EutS, GLYCEROL, POTASSIUM ION, ... | | Authors: | Ochoa, J.M, Sawaya, M.R, Nguyen, V.N, Duilio, C, Yeates, T.O, Nie, M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Symmetry breaking and structural polymorphism in a bacterial microcompartment shell protein for choline utilization.
Protein Sci., 29, 2020
|
|
6XPL
 
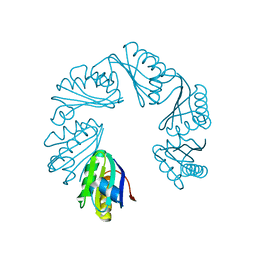 | | CutR Screw, form 2 with 33.8 angstrom pitch | | Descriptor: | Ethanolamine utilization protein EutS | | Authors: | Ochoa, J.M, Sawaya, M.R, Nguyen, V.N, Duilio, C, Yeates, T.O. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Symmetry breaking and structural polymorphism in a bacterial microcompartment shell protein for choline utilization.
Protein Sci., 29, 2020
|
|
6XPI
 
 | | CutR flat hexamer, form 1 | | Descriptor: | Ethanolamine utilization protein EutS | | Authors: | Ochoa, J.M, Sawaya, M.R, Nguyen, V.N, Duilio, C, Yeates, T.O. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Symmetry breaking and structural polymorphism in a bacterial microcompartment shell protein for choline utilization.
Protein Sci., 29, 2020
|
|
6XPJ
 
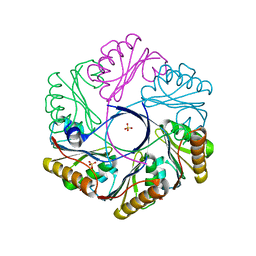 | | CutR flat hexamer, form 2 | | Descriptor: | Ethanolamine utilization protein EutS, SULFATE ION | | Authors: | Ochoa, J.M, Sawaya, M.R, Nguyen, V.N, Duilio, C, Yeates, T.O. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Symmetry breaking and structural polymorphism in a bacterial microcompartment shell protein for choline utilization.
Protein Sci., 29, 2020
|
|
6WQK
 
 | | hnRNPA2 Low complexity domain (LCD) determined by cryoEM | | Descriptor: | MCherry fluorescent protein,Heterogeneous nuclear ribonucleoproteins A2/B1 chimera | | Authors: | Lu, J, Cao, Q, Hughes, M.P, Sawaya, M.R, Boyer, D.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2020-04-29 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | CryoEM structure of the low-complexity domain of hnRNPA2 and its conversion to pathogenic amyloid.
Nat Commun, 11, 2020
|
|
7R1E
 
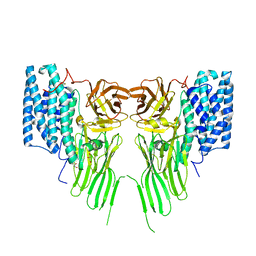 | | Mosquitocidal Cry11Ba determined at pH 10.4 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | GLYCEROL, Pesticidal crystal protein Cry11Ba | | Authors: | Colletier, J.-P, Sawaya, M.R, Schibrowsky, N.A, Cascio, D, Rodriguez, J.A. | | Deposit date: | 2022-02-02 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
6XPK
 
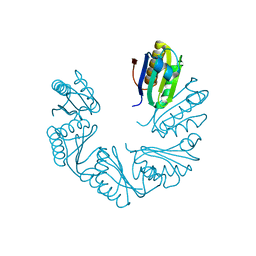 | | CutR Screw, form 1 with 41.9 angstrom pitch | | Descriptor: | Ethanolamine utilization protein EutS | | Authors: | Ochoa, J.M, Sawaya, M.R, Nguyen, V.N, Duilio, C, Yeates, T.O. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Symmetry breaking and structural polymorphism in a bacterial microcompartment shell protein for choline utilization.
Protein Sci., 29, 2020
|
|
7SAS
 
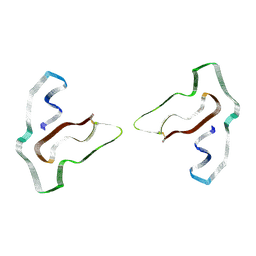 | | Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Cao, Q, Jiang, Y, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-09-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Amyloid fibrils in FTLD-TDP are composed of TMEM106B and not TDP-43.
Nature, 605, 2022
|
|
7SAQ
 
 | | Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Cao, Q, Jiang, Y, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-09-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Amyloid fibrils in FTLD-TDP are composed of TMEM106B and not TDP-43.
Nature, 605, 2022
|
|
7SAR
 
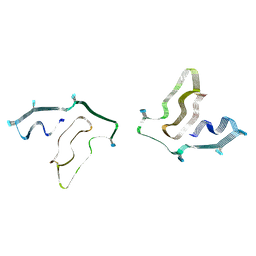 | | Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Cao, Q, Jiang, Y, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-09-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Amyloid fibrils in FTLD-TDP are composed of TMEM106B and not TDP-43.
Nature, 605, 2022
|
|
1NOM
 
 | |
6ODG
 
 | | SVQIVY, Crystal Structure of a tau protein fragment | | Descriptor: | Microtubule-associated protein tau | | Authors: | Eisenberg, D.S, Boyer, D.R, Sawaya, M.R, Seidler, P.M. | | Deposit date: | 2019-03-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure-based inhibitors halt prion-like seeding by Alzheimer's disease-and tauopathy-derived brain tissue samples.
J.Biol.Chem., 294, 2019
|
|
6PES
 
 | | Cryo-EM structure of alpha-synuclein H50Q Wide Fibril | | Descriptor: | Alpha-synuclein | | Authors: | Boyer, D.R, Li, B, Sawaya, M.R, Jiang, L, Eisenberg, D.S. | | Deposit date: | 2019-06-20 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of fibrils formed by alpha-synuclein hereditary disease mutant H50Q reveal new polymorphs.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6PIL
 
 | | Antibody scFv-M204 monomeric state | | Descriptor: | CHLORIDE ION, scFv-M204 antibody | | Authors: | Abskharon, R, Sawaya, M.R, Seidler, P.M, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2019-06-26 | | Release date: | 2020-06-24 | | Last modified: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a conformational antibody that binds tau oligomers and inhibits pathological seeding by extracts from donors with Alzheimer's disease.
J.Biol.Chem., 295, 2020
|
|
6O4J
 
 | | Amyloid Beta KLVFFAENVGS 16-26 D23N Iowa mutation | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Griner, S.L, Sawaya, M.R, Rodriguez, J.A, Cascio, D, Gonen, T. | | Deposit date: | 2019-02-28 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.402 Å) | | Cite: | Structure based inhibitors of Amyloid Beta core suggest a common interface with Tau.
Elife, 8, 2019
|
|
