7DAT
 
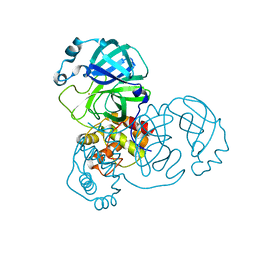 | | The crystal structure of COVID-19 main protease treated by AF | | Descriptor: | COVID-19 MAIN PROTEASE, GOLD ION | | Authors: | He, Z.S, He, B, Cao, P, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2020-10-18 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A comparison of Remdesivir versus gold cluster in COVID-19 animal model: A better therapeutic outcome of gold cluster.
Nano Today, 44, 2022
|
|
7DAV
 
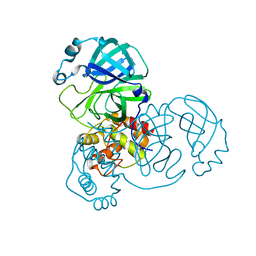 | | The native crystal structure of COVID-19 main protease | | Descriptor: | COVID-19 MAIN PROTEASE | | Authors: | He, Z.S, He, B, Cao, P, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2020-10-18 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A comparison of Remdesivir versus gold cluster in COVID-19 animal model: A better therapeutic outcome of gold cluster.
Nano Today, 44, 2022
|
|
7DAU
 
 | | The crystal structure of COVID-19 main protease treated by GA | | Descriptor: | COVID-19 MAIN PROTEASE, GOLD ION | | Authors: | He, Z.S, He, B, Cao, P, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2020-10-18 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A comparison of Remdesivir versus gold cluster in COVID-19 animal model: A better therapeutic outcome of gold cluster.
Nano Today, 44, 2022
|
|
7B3Y
 
 | | Structure of a nanoparticle for a COVID-19 vaccine candidate | | Descriptor: | Fibronectin binding protein,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase | | Authors: | Duyvesteyn, H.M.E, Stuart, D.I. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses.
Nat Commun, 12, 2021
|
|
7RKU
 
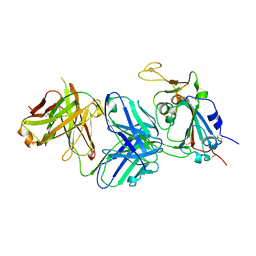 | | Structure of the SARS-CoV-2 receptor binding domain in complex with the human neutralizing antibody Fab fragment, C022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C022 Antibody Fab Heavy Chain, C022 Antibody Fab Light Chain, ... | | Authors: | Jette, C.A, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
6Y84
 
 | | SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19) | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Owen, C.D, Lukacik, P, Strain-Damerell, C.M, Douangamath, A, Powell, A.J, Fearon, D, Brandao-Neto, J, Crawshaw, A.D, Aragao, D, Williams, M, Flaig, R, Hall, D.R, McAuley, K.E, Mazzorana, M, Stuart, D.I, von Delft, F, Walsh, M.A. | | Deposit date: | 2020-03-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | COVID-19 main protease with unliganded active site
To Be Published
|
|
6YB7
 
 | | SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19). | | Descriptor: | 3C-like proteinase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE | | Authors: | Owen, C.D, Lukacik, P, Strain-Damerell, C.M, Douangamath, A, Powell, A.J, Fearon, D, Brandao-Neto, J, Crawshaw, A.D, Aragao, D, Williams, M, Flaig, R, Hall, D.R, McAuley, K.E, Mazzorana, M, Stuart, D.I, von Delft, F, Walsh, M.A. | | Deposit date: | 2020-03-16 | | Release date: | 2020-03-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | COVID-19 main protease with unliganded active site
To Be Published
|
|
7WKJ
 
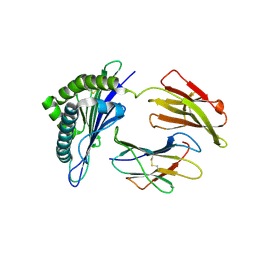 | | A COVID-19 T-cell response detection method based on a newly identified human CD8+ T cell epitope from SARS-CoV-2-Hubei Province, 2021. | | Descriptor: | Beta-2-microglobulin, LYS-THR-PHE-PRO-PRO-THR-GLU-PRO-LYS, MHC class I antigen | | Authors: | Zhang, J, Lu, D, Li, M, Liu, M.S, Yao, S.J, Zhan, J.B, Liu, J, Gao, G.F. | | Deposit date: | 2022-01-10 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A COVID-19 T-Cell Response Detection Method Based on a Newly Identified Human CD8 + T Cell Epitope from SARS-CoV-2 - Hubei Province, China, 2021.
China CDC Wkly, 4, 2022
|
|
7BZ5
 
 | | Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of B38, Light chain of B38, ... | | Authors: | Wu, Y, Qi, J, Gao, F. | | Deposit date: | 2020-04-26 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2.
Science, 368, 2020
|
|
7M3I
 
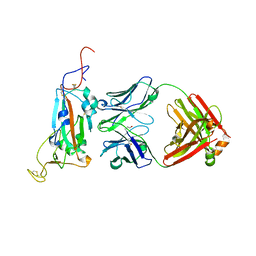 | |
6W63
 
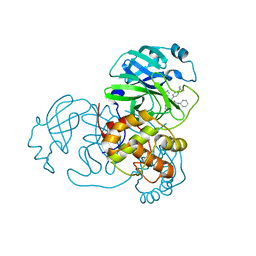 | |
6W4B
 
 | | The crystal structure of Nsp9 RNA binding protein of SARS CoV-2 | | Descriptor: | Non-structural protein 9 | | Authors: | Tan, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-10 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of Nsp9 replicase protein of COVID-19
To Be Published
|
|
6M71
 
 | | SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-01 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
7B3O
 
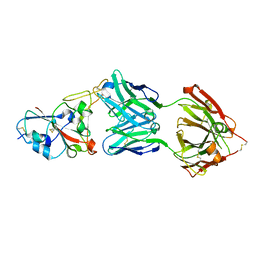 | | Crystal structure of the SARS-CoV-2 RBD in complex with STE90-C11 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Fab Fragment, Light Chain of Fab Fragment, ... | | Authors: | Kluenemann, T, Van den Heuvel, J. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A SARS-CoV-2 neutralizing antibody selected from COVID-19 patients binds to the ACE2-RBD interface and is tolerant to most known RBD mutations.
Cell Rep, 36, 2021
|
|
6LU7
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor N3 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | Deposit date: | 2020-01-26 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
7OAO
 
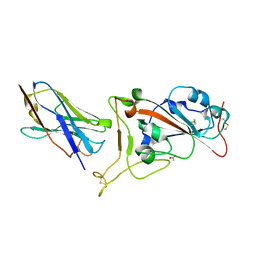 | | Nanobody C5 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, C5 nanobody, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAQ
 
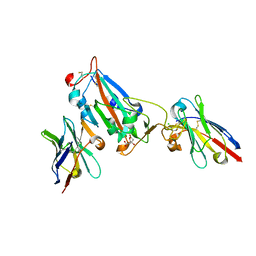 | | Nanobody H3 AND C1 bound to RBD with Kent mutation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAP
 
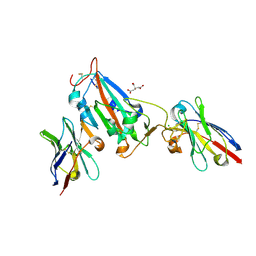 | | Nanobody H3 AND C1 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C1 nanobody, CHLORIDE ION, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAN
 
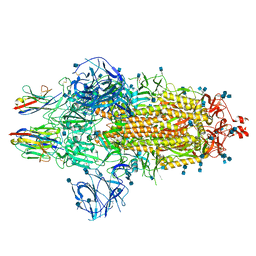 | | Nanobody C5 bound to Spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Naismith, J.H, Weckener, M. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-11 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAY
 
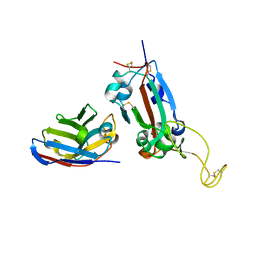 | | Nanobody F2 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2 nanobody, Spike protein S1 | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAU
 
 | | Nanobody C5 bound to Kent variant RBD (N501Y) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C5, GLYCEROL, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7AEH
 
 | | SARS-CoV-2 main protease in a covalent complex with a pyridine derivative of ABT-957, compound 1 | | Descriptor: | (2~{R})-5-oxidanylidene-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]-1-(phenylmethyl)pyrrolidine-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Owen, C.D, Redhead, M.A, Lukacik, P, Strain-Damerell, C, Fearon, D, Brewitz, L, Collette, A, Robinson, C, Collins, P, Radoux, C, Navratilova, I, Douangamath, A, von Delft, F, Malla, T.R, Nugen, T, Hull, H, Tumber, A, Schofield, C.J, Hallet, D, Stuart, D.I, Hopkins, A.L, Walsh, M.A. | | Deposit date: | 2020-09-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Bispecific repurposed medicines targeting the viral and immunological arms of COVID-19.
Sci Rep, 11, 2021
|
|
8P2Y
 
 | | Structure of human SIT1:ACE2 complex (closed PD conformation) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P2X
 
 | | Structure of human SIT1:ACE2 complex (open PD conformation) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P2W
 
 | | Structure of human SIT1 (focussed map / refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Sodium- and chloride-dependent transporter XTRP3 | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
