8ES6
 
 | |
6YHV
 
 | | Structural insights into Pseudomonas aeruginosa Type six secretion system exported effector 8: unliganded Tse8 | | Descriptor: | COPPER (II) ION, Tse8 | | Authors: | Sainz-Polo, M.A, Capuni, R, Pretre, G, Gonzalez-Magana, A, Lucas, M, Altuna, J, Montanchez, I, Fucini, P, Albesa-Jove, D. | | Deposit date: | 2020-03-31 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Structural insights into Pseudomonas aeruginosaType six secretion system exported effector 8.
J.Struct.Biol., 212, 2020
|
|
6TE4
 
 | | Structural insights into Pseudomonas aeruginosa Type six secretion system exported effector 8: Tse8 in complex with a peptide | | Descriptor: | Pro-Pro-Leu-Ala-Ser-Lys, Tse8 | | Authors: | Sainz-Polo, M.A, Capuni, R, Lucas, M, Altuna, J, Fucini, P, Montanchez, I, Albesa-Jove, D. | | Deposit date: | 2019-11-11 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural insights into Pseudomonas aeruginosaType six secretion system exported effector 8.
J.Struct.Biol., 212, 2020
|
|
5AC3
 
 | | Crystal structure of PAM12A | | Descriptor: | ACETIC ACID, CADMIUM ION, PEPTIDE AMIDASE | | Authors: | Wu, B, Wijma, H.J, Song, L, Rozeboom, H.J, Poloni, C, Tian, Y, Arif, M.I, Nuijens, T, Quadflieg, P.J.L.M, Szymanski, W, Feringa, B.L, Janssen, D.B. | | Deposit date: | 2015-08-11 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Versatile Peptide C-Terminal Functionalization Via a Computationally Peptide Amidase
Acs Catalysis, 2016
|
|
6DII
 
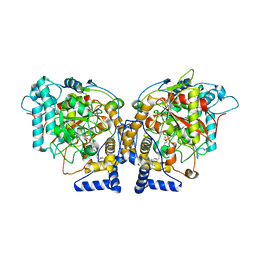 | | Structure of Arabidopsis Fatty Acid Amide Hydrolase in Complex with methyl linolenyl fluorophosphonate | | Descriptor: | Fatty acid amide hydrolase, methyl-9Z,12Z,15Z-octadecatrienylphosphonofluoridate | | Authors: | Aziz, M, Wang, X, Tripathi, A, Bankaitis, V, Chapman, K.D. | | Deposit date: | 2018-05-23 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural analysis of a plant fatty acid amide hydrolase provides insights into the evolutionary diversity of bioactive acylethanolamides.
J.Biol.Chem., 294, 2019
|
|
6DHV
 
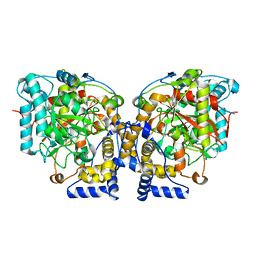 | | Structure of Arabidopsis Fatty Acid Amide Hydrolase | | Descriptor: | Fatty acid amide hydrolase | | Authors: | Aziz, M, Wang, X, Tripathi, A, Bankaitis, V, Chapman, K.D. | | Deposit date: | 2018-05-21 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural analysis of a plant fatty acid amide hydrolase provides insights into the evolutionary diversity of bioactive acylethanolamides.
J.Biol.Chem., 294, 2019
|
|
4YJI
 
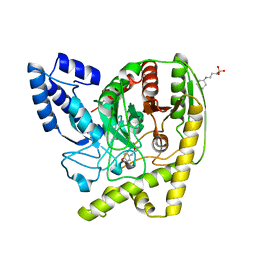 | | The Crystal Structure of a Bacterial Aryl Acylamidase Belonging to the Amidase signature (AS) enzymes family | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Aryl acylamidase, N-(4-HYDROXYPHENYL)ACETAMIDE (TYLENOL) | | Authors: | Choi, I.-G, Lee, S, Park, E.-H, Ko, H.-J, Bang, W.-G. | | Deposit date: | 2015-03-03 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure analysis of a bacterial aryl acylamidase belonging to the amidase signature enzyme family
Biochem.Biophys.Res.Commun., 467, 2015
|
|
4YJ6
 
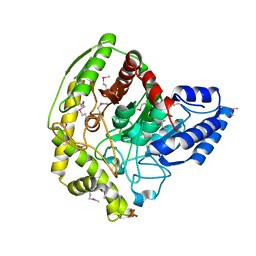 | | The Crystal Structure of a Bacterial Aryl Acylamidase Belonging to the Amidase signature (AS) enzymes family | | Descriptor: | Aryl acylamidase, PHOSPHATE ION | | Authors: | Lee, S, Park, E.-H, Ko, H.-J, Bang, W.-G, Choi, I.-G. | | Deposit date: | 2015-03-03 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure analysis of a bacterial aryl acylamidase belonging to the amidase signature enzyme family
Biochem.Biophys.Res.Commun., 467, 2015
|
|
4CP8
 
 | | Structure of the amidase domain of allophanate hydrolase from Pseudomonas sp strain ADP | | Descriptor: | ALLOPHANATE HYDROLASE, MALONATE ION | | Authors: | Balotra, S, Newman, J, French, N, French, L, Peat, T.S, Scott, C. | | Deposit date: | 2014-02-03 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray Structure of the Amidase Domain of Atzf, the Allophanate Hydrolase from the Cyanuric Acid-Mineralizing Multienzyme Complex.
Appl.Environ.Microbiol., 81, 2015
|
|
6KVR
 
 | | Fatty acid amide hydrolase | | Descriptor: | Fatty acid amide hydrolase | | Authors: | Min, C.A, Yun, J.S, Chang, J.H. | | Deposit date: | 2019-09-05 | | Release date: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparison of Candida Albicans Fatty Acid Amide Hydrolase Structure with Homologous Amidase Signature Family Enzymes
Crystals, 9, 2019
|
|
4DO3
 
 | | Structure of FAAH with a non-steroidal anti-inflammatory drug | | Descriptor: | (2S)-2-(6-chloro-9H-carbazol-2-yl)propanoic acid, CHLORIDE ION, CYCLOHEXANE AMINOCARBOXYLIC ACID, ... | | Authors: | Garau, G. | | Deposit date: | 2012-02-09 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Binding Site for Nonsteroidal Anti-inflammatory Drugs in Fatty Acid Amide Hydrolase.
J.Am.Chem.Soc., 135, 2013
|
|
5EWQ
 
 | | The crystal structure of an amidase family protein from Bacillus anthracis str. Ames | | Descriptor: | ACETATE ION, Amidase | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-20 | | Release date: | 2015-12-09 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The crystal structure of an amidase family protein from Bacillus anthracis str. Ames
To Be Published
|
|
6MRG
 
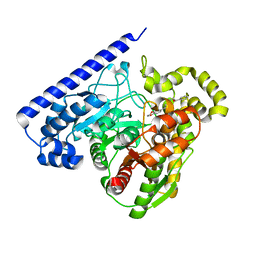 | | FAAH bound to non covalent inhibitor | | Descriptor: | (1R)-2-{[6-(2,3-dihydro-1,4-benzodioxin-6-yl)pyrimidin-4-yl]amino}-1-phenylethan-1-ol, Fatty-acid amide hydrolase 1 | | Authors: | Saha, A, Shih, A, Mirzadegan, T, Seierstad, M. | | Deposit date: | 2018-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Predicting the Binding of Fatty Acid Amide Hydrolase Inhibitors by Free Energy Perturbation.
J Chem Theory Comput, 14, 2018
|
|
1M22
 
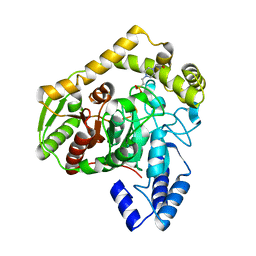 | | X-ray structure of native peptide amidase from Stenotrophomonas maltophilia at 1.4 A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, peptide amidase | | Authors: | Labahn, J, Neumann, S, Buldt, G, Kula, M.-R, Granzin, J. | | Deposit date: | 2002-06-21 | | Release date: | 2002-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An alternative mechanism for amidase signature enzymes
J.MOL.BIOL., 322, 2002
|
|
1M21
 
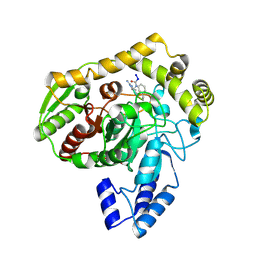 | | Crystal structure analysis of the peptide amidase PAM in complex with the competitive inhibitor chymostatin | | Descriptor: | CHYMOSTATIN, Peptide Amidase | | Authors: | Labahn, J, Neumann, S, Buldt, G, Kula, M.-R, Granzin, J. | | Deposit date: | 2002-06-21 | | Release date: | 2002-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An alternative mechanism for amidase signature enzymes
J.MOL.BIOL., 322, 2002
|
|
2DC0
 
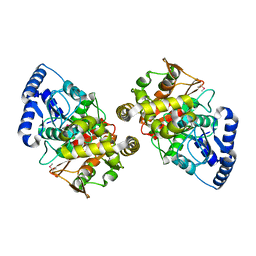 | | Crystal structure of amidase | | Descriptor: | probable amidase | | Authors: | Ohshima, T, Sakuraba, H, Ebihara, A, Kanagawa, M, Nakagawa, N, Kuroishi, C, Satoh, S, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-17 | | Release date: | 2007-01-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of amidase
To be Published
|
|
2VYA
 
 | | Crystal Structure of fatty acid amide hydrolase conjugated with the drug-like inhibitor PF-750 | | Descriptor: | 4-(quinolin-3-ylmethyl)piperidine-1-carboxylic acid, CHLORIDE ION, FATTY-ACID AMIDE HYDROLASE 1, ... | | Authors: | Mileni, M, Johnson, D.S, Wang, Z, Everdeen, D.S, Liimatta, M, Pabst, B, Bhattacharya, K, Nugent, R.A, Kamtekar, S, Cravatt, B.F, Ahn, K, Stevens, R.C. | | Deposit date: | 2008-07-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Guided Inhibitor Design for Human Faah by Interspecies Active Site Conversion.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2WAP
 
 | | 3D-crystal structure of humanized-rat fatty acid amide hydrolase (FAAH) conjugated with the drug-like urea inhibitor PF-3845 | | Descriptor: | 4-(3-{[5-(trifluoromethyl)pyridin-2-yl]oxy}benzyl)piperidine-1-carboxylic acid, CHLORIDE ION, FATTY-ACID AMIDE HYDROLASE 1, ... | | Authors: | Mileni, M, Kamtekar, S, Stevens, R.C. | | Deposit date: | 2009-02-11 | | Release date: | 2009-05-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Characterization of a Highly Selective Faah Inhibitor that Reduces Inflammatory Pain.
Chem.Biol., 16, 2009
|
|
2GI3
 
 | |
2WJ1
 
 | | 3D-crystal structure of humanized-rat fatty acid amide hydrolase (FAAH) conjugated with 7-phenyl-1-(4-(pyridin-2-yl)oxazol-2-yl)heptan- 1-one, an alpha-ketooxazole | | Descriptor: | 7-phenyl-1-(4-pyridin-2-yl-1,3-oxazol-2-yl)heptane-1,1-diol, CHLORIDE ION, FATTY-ACID AMIDE HYDROLASE 1 | | Authors: | Mileni, M, Garfunkle, J, DeMartino, J.K, Cravatt, B.F, Boger, D.L, Stevens, R.C. | | Deposit date: | 2009-05-19 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Binding and Inactivation Mechanism of a Humanized Fatty Acid Amide Hydrolase by Alpha-Ketoheterocycle Inhibitors Revealed from Cocrystal Structures.
J.Am.Chem.Soc., 131, 2009
|
|
2WJ2
 
 | | 3D-crystal structure of humanized-rat fatty acid amide hydrolase (FAAH) conjugated with 7-phenyl-1-(5-(pyridin-2-yl)oxazol-2-yl)heptan- 1-one, an alpha-ketooxazole | | Descriptor: | 7-phenyl-1-(5-pyridin-2-yl-1,3-oxazol-2-yl)heptane-1,1-diol, CHLORIDE ION, FATTY ACID AMIDE HYDROLASE 1 | | Authors: | Mileni, M, Garfunkle, J, DeMartino, J.K, Cravatt, B.F, Boger, D.L, Stevens, R.C. | | Deposit date: | 2009-05-19 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Binding and Inactivation Mechanism of a Humanized Fatty Acid Amide Hydrolase by Alpha-Ketoheterocycle Inhibitors Revealed from Cocrystal Structures.
J.Am.Chem.Soc., 131, 2009
|
|
3A1K
 
 | | Crystal structure of Rhodococcus sp. N771 Amidase | | Descriptor: | Amidase | | Authors: | Ohtaki, A, Noguchi, K, Sato, Y, Murata, K, Odaka, M, Yohda, M. | | Deposit date: | 2009-04-09 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and characterization of amidase from Rhodococcus sp. N-771: Insight into the molecular mechanism of substrate recognition
Biochim.Biophys.Acta, 1804, 2010
|
|
3A2Q
 
 | | Structure of 6-aminohexanoate cyclic dimer hydrolase complexed with substrate | | Descriptor: | 6-AMINOHEXANOIC ACID, 6-aminohexanoate-cyclic-dimer hydrolase, GLYCEROL | | Authors: | Shibata, N. | | Deposit date: | 2009-05-26 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic analysis of the 6-aminohexanoate cyclic dimer hydrolase: catalytic mechanism and evolution of an enzyme responsible for nylon-6 byproduct degradation
J.Biol.Chem., 285, 2010
|
|
3A2P
 
 | | Structure of 6-aminohexanoate cyclic dimer hydrolase | | Descriptor: | 6-aminohexanoate-cyclic-dimer hydrolase, GLYCEROL | | Authors: | Shibata, N. | | Deposit date: | 2009-05-26 | | Release date: | 2009-11-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystallographic analysis of the 6-aminohexanoate cyclic dimer hydrolase: catalytic mechanism and evolution of an enzyme responsible for nylon-6 byproduct degradation
J.Biol.Chem., 285, 2010
|
|
3A1I
 
 | | Crystal structure of Rhodococcus sp. N-771 Amidase complexed with Benzamide | | Descriptor: | Amidase, BENZAMIDE | | Authors: | Ohtaki, A, Noguchi, K, Sato, Y, Murata, K, Odaka, M, Yohda, M. | | Deposit date: | 2009-04-03 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and Characterization of Amidase from Rhodococcus sp. N-771: Insight into the Molecular Mechanism of Substrate Recognition
Biochim.Biophys.Acta, 2009
|
|
