1WE9
 
 | | Solution structure of PHD domain in nucleic acid binding protein-like NP_197993 | | Descriptor: | PHD finger family protein, ZINC ION | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-24 | | Release date: | 2004-11-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PHD domain in nucleic acid binding protein-like NP_197993
To be Published
|
|
1WEN
 
 | | Solution structure of PHD domain in ING1-like protein BAC25079 | | Descriptor: | ZINC ION, inhibitor of growth family, member 4; ING1-like protein | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2004-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PHD domain in ING1-like protein BAC25079
To be Published
|
|
1WES
 
 | | Solution structure of PHD domain in inhibitor of growth family, member 1-like | | Descriptor: | ZINC ION, inhibitor of growth family, member 1-like | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2004-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PHD domain in inhibitor of growth family, member 1-like
To be Published
|
|
1WEU
 
 | | Solution structure of PHD domain in ING1-like protein BAC25009 | | Descriptor: | ZINC ION, inhibitor of growth family, member 4 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2004-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PHD domain in ING1-like protein BAC25009
To be Published
|
|
1X4I
 
 | | Solution structure of PHD domain in inhibitor of growth protein 3 (ING3) | | Descriptor: | Inhibitor of growth protein 3, ZINC ION | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PHD domain in inhibitor of growth protein 3 (ING3)
To be Published
|
|
2F6J
 
 | |
2F6N
 
 | |
2FSA
 
 | |
2FUI
 
 | |
2FUU
 
 | |
2G6Q
 
 | |
2GF7
 
 | | Double tudor domain structure | | Descriptor: | Jumonji domain-containing protein 2A, SULFATE ION | | Authors: | Huang, Y, Fang, J, Bedford, M.T, Zhang, Y, Xu, R.M. | | Deposit date: | 2006-03-21 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A
Science, 312, 2006
|
|
2GFA
 
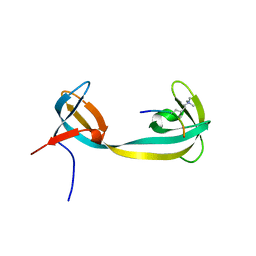 | | double tudor domain complex structure | | Descriptor: | Jumonji domain-containing protein 2A, peptide | | Authors: | Huang, Y, Fang, J, Bedford, M.T, Zhang, Y, Xu, R.M. | | Deposit date: | 2006-03-21 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A
Science, 312, 2006
|
|
2JMI
 
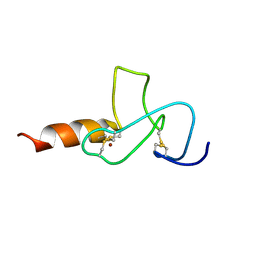 | | NMR solution structure of PHD finger fragment of Yeast Yng1 protein in free state | | Descriptor: | Protein YNG1, ZINC ION | | Authors: | Ilin, S, Taverna, S.D, Rogers, R.S, Tanny, J.C, Lavender, H, Li, H, Baker, L, Boyle, J, Blair, L.P, Chait, B.T, Patel, D.J, Aitchison, J.D, Tackett, A.J, Allis, C.D. | | Deposit date: | 2006-11-15 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs
Mol.Cell, 24, 2006
|
|
2JMJ
 
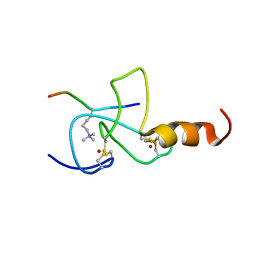 | | NMR solution structure of the PHD domain from the yeast YNG1 protein in complex with H3(1-9)K4me3 peptide | | Descriptor: | Histone H3, Protein YNG1, ZINC ION | | Authors: | Ilin, S, Taverna, S.D, Rogers, R.S, Tanny, J.C, Lavender, H, Li, H, Baker, L, Boyle, J, Blair, L.P, Chait, B.T, Patel, D.J, Aitchison, J.D, Tackett, A.J, Allis, C.D. | | Deposit date: | 2006-11-15 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs
Mol.Cell, 24, 2006
|
|
2K16
 
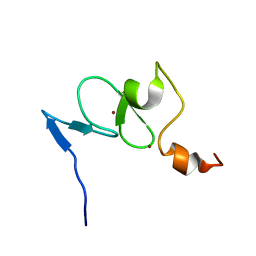 | | Solution structure of the free TAF3 PHD domain | | Descriptor: | Transcription initiation factor TFIID subunit 3, ZINC ION | | Authors: | van Ingen, H, van Schaik, F.M.A, Wienk, H, Timmers, M, Boelens, R. | | Deposit date: | 2008-02-22 | | Release date: | 2008-07-15 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Recognition of the H3K4me3 mark by the TAF3-PHD finger
To be Published
|
|
2K17
 
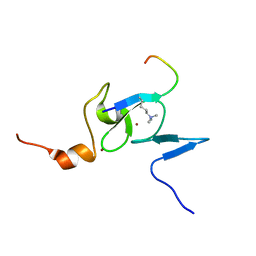 | | Solution structure of the TAF3 PHD domain in complex with a H3K4me3 peptide | | Descriptor: | H3K4me3 peptide, Transcription initiation factor TFIID subunit 3, ZINC ION | | Authors: | van Ingen, H, van Schaik, F.M.A, Wienk, H, Timmers, M, Boelens, R. | | Deposit date: | 2008-02-22 | | Release date: | 2008-07-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Recognition of the H3K4me3 mark by the TAF3-PHD finger
To be Published
|
|
2K1J
 
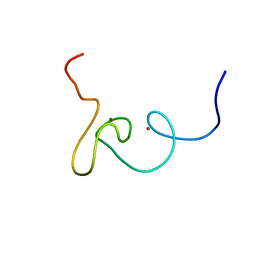 | | Plan homeodomain finger of tumour supressor ING4 | | Descriptor: | Inhibitor of growth protein 4, ZINC ION | | Authors: | Palacios, A, Garcia, P, Padro, D, Lopez-Hernandez, E, Blanco, F.J. | | Deposit date: | 2008-03-05 | | Release date: | 2008-04-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and NMR characterization of the binding to methylated histone tails of the plant homeodomain finger of the tumour suppressor ING4.
Febs Lett., 580, 2006
|
|
2KGG
 
 | |
2KGI
 
 | |
2KU7
 
 | |
2KYU
 
 | | The solution structure of the PHD3 finger of MLL | | Descriptor: | Histone-lysine N-methyltransferase MLL, ZINC ION | | Authors: | Park, S, Bushweller, J.H. | | Deposit date: | 2010-06-08 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The PHD3 domain of MLL acts as a CYP33-regulated switch between MLL-mediated activation and repression.
Biochemistry, 49, 2010
|
|
2KYX
 
 | | Solution structure of the RRM domain of CYP33 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E | | Authors: | Park, S, Bushweller, J.H. | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The PHD3 domain of MLL acts as a CYP33-regulated switch between MLL-mediated activation and repression .
Biochemistry, 49, 2010
|
|
2M1R
 
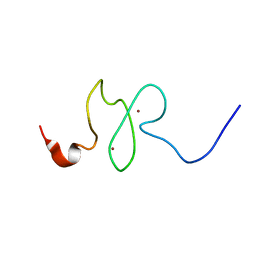 | | PHD domain of ING4 N214D mutant | | Descriptor: | Inhibitor of growth protein 4, ZINC ION | | Authors: | Blanco, F.J. | | Deposit date: | 2012-12-05 | | Release date: | 2012-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Functional impact of cancer-associated mutations in the tumor suppressor protein ING4.
Carcinogenesis, 31, 2010
|
|
2M3H
 
 | |
