5FF8
 
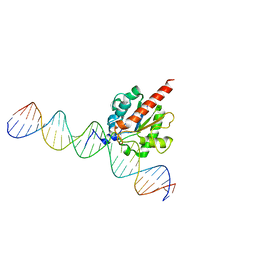 | | TDG enzyme-product complex | | Descriptor: | DNA, G/T mismatch-specific thymine DNA glycosylase | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2015-12-18 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues.
Nucleic Acids Res., 44, 2016
|
|
4UQM
 
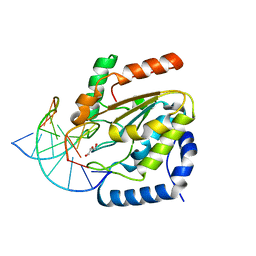 | | Crystal structure determination of uracil-DNA N-glycosylase (UNG) from Deinococcus radiodurans in complex with DNA - new insights into the role of the Leucine-loop for damage recognition and repair | | Descriptor: | 5'-D(*CP*CP*TP*AP*TP*CP*CP*AP*AAB*GP*TP*CP*TP*CP*CP*G)-3', 5'-D(*GP*CP*GP*GP*AP*GP*AP*CP*AP*TP*GP*GP*AP*CP*AP*G)-3', CHLORIDE ION, ... | | Authors: | Pedersen, H.L, Johnson, K.A, McVey, C.E, Leiros, I, Moe, E. | | Deposit date: | 2014-06-24 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure determination of uracil-DNA N-glycosylase from Deinococcus radiodurans in complex with DNA.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
1AKZ
 
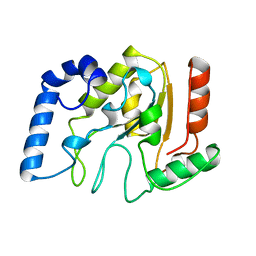 | | HUMAN URACIL-DNA GLYCOSYLASE | | Descriptor: | URACIL-DNA GLYCOSYLASE | | Authors: | Tainer, J.A, Mol, C.D. | | Deposit date: | 1997-05-27 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
Embo J., 17, 1998
|
|
3IKB
 
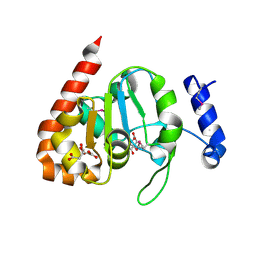 | |
5JXY
 
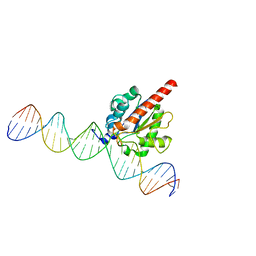 | | Enzyme-substrate complex of TDG catalytic domain bound to a G/U analog | | Descriptor: | DNA (28-MER), G/T mismatch-specific thymine DNA glycosylase | | Authors: | Pidugu, L.S, Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2016-05-13 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues.
Nucleic Acids Res., 44, 2016
|
|
4SKN
 
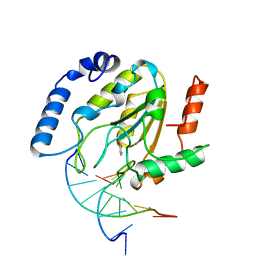 | | A NUCLEOTIDE-FLIPPING MECHANISM FROM THE STRUCTURE OF HUMAN URACIL-DNA GLYCOSYLASE BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*CP*CP*GP*CP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*GP*(D1P)P*GP*GP*CP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE), ... | | Authors: | Slupphaug, G, Mol, C.D, Kavli, B, Arvai, A.S, Krokan, H.E, Tainer, J.A. | | Deposit date: | 1999-02-20 | | Release date: | 1999-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA.
Nature, 384, 1996
|
|
5T2W
 
 | |
3A7N
 
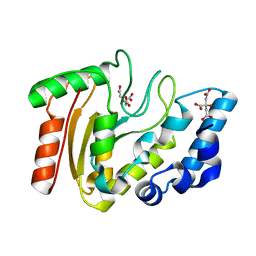 | | Crystal structure of uracil-DNA glycosylase from Mycobacterium tuberculosis | | Descriptor: | CITRATE ANION, Uracil-DNA glycosylase | | Authors: | Kaushal, P.S, Talawar, R.K, Varshney, U, Vijayan, M. | | Deposit date: | 2009-09-29 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of uracil-DNA glycosylase from Mycobacterium tuberculosis: insights into interactions with ligands
Acta Crystallogr.,Sect.F, 66, 2010
|
|
4JGC
 
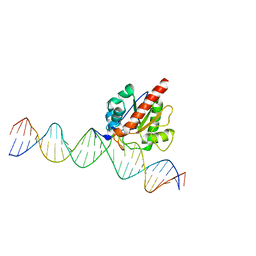 | | Human TDG N140A mutant IN A COMPLEX WITH 5-carboxylcytosine (5caC) | | Descriptor: | 4-amino-2-oxo-1,2-dihydropyrimidine-5-carboxylic acid, G/T mismatch-specific thymine DNA glycosylase, oligonucleotide, ... | | Authors: | Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2013-02-28 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.582 Å) | | Cite: | Activity and crystal structure of human thymine DNA glycosylase mutant N140A with 5-carboxylcytosine DNA at low pH.
Dna Repair, 12, 2013
|
|
5EUG
 
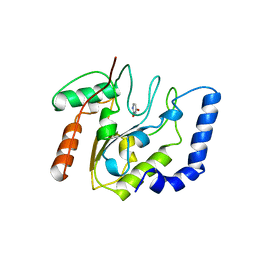 | | CRYSTALLOGRAPHIC AND ENZYMATIC STUDIES OF AN ACTIVE SITE VARIANT H187Q OF ESCHERICHIA COLI URACIL DNA GLYCOSYLASE: CRYSTAL STRUCTURES OF MUTANT H187Q AND ITS URACIL COMPLEX | | Descriptor: | PROTEIN (GLYCOSYLASE), URACIL | | Authors: | Xiao, G, Tordova, M, Drohat, A.C, Jagadeesh, J, Stivers, J.T, Gilliland, G.L. | | Deposit date: | 1998-12-27 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited.
Proteins, 35, 1999
|
|
5CYS
 
 | |
4EUG
 
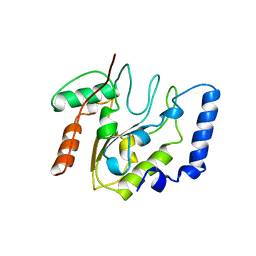 | | Crystallographic and Enzymatic Studies of an Active Site Variant H187Q of Escherichia Coli Uracil DNA Glycosylase: Crystal Structures of Mutant H187Q and its Uracil Complex | | Descriptor: | PROTEIN (GLYCOSYLASE) | | Authors: | Xiao, G, Tordova, M, Drohat, A.C, Jagadeesh, J, Stivers, J.T, Gilliland, G.L. | | Deposit date: | 1998-12-27 | | Release date: | 1999-07-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Heteronuclear NMR and crystallographic studies of wild-type and H187Q Escherichia coli uracil DNA glycosylase: electrophilic catalysis of uracil expulsion by a neutral histidine 187.
Biochemistry, 38, 1999
|
|
4FNC
 
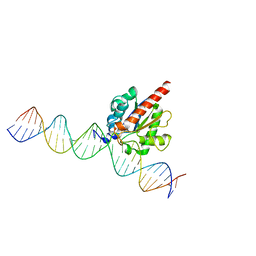 | | Human TDG in a post-reactive complex with 5-hydroxymethyluracil (5hmU) | | Descriptor: | 5-HYDROXYMETHYL URACIL, DNA (28-MER), DNA (29-MER), ... | | Authors: | Hashimoto, H, Hong, S, Bhagwat, A.S, Zhang, X, Cheng, X. | | Deposit date: | 2012-06-19 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: its structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
2SSP
 
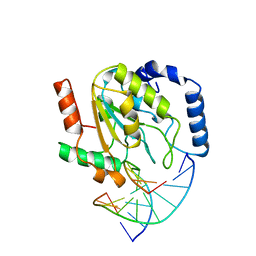 | | LEUCINE-272-ALANINE URACIL-DNA GLYCOSYLASE BOUND TO ABASIC SITE-CONTAINING DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*TP*(AAB)P*AP*TP*CP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE) | | Authors: | Parikh, S.S, Mol, C.D, Slupphaug, G, Bharati, S, Krokan, H.E, Tainer, J.A. | | Deposit date: | 1999-04-28 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
EMBO J., 17, 1998
|
|
1VK2
 
 | |
1YUO
 
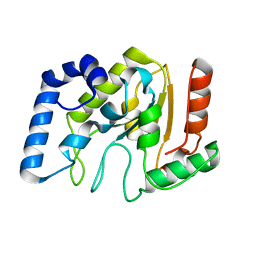 | | Optimisation of the surface electrostatics as a strategy for cold adaptation of uracil-DNA N-glycosylase (UNG)from atlantic cod (Gadus morhua) | | Descriptor: | Uracil-DNA glycosylase | | Authors: | Moe, E, Leiros, I, Riise, E.K, Olufsen, M, Lanes, O, Smalas, A.O, Willassen, N.P. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Optimisation of the surface electrostatics as a strategy for cold adaptation of uracil-DNA N-glycosylase (UNG) from Atlantic cod (Gadus morhua)
J.Mol.Biol., 343, 2004
|
|
5NNH
 
 | | KSHV uracil-DNA glycosylase, apo form | | Descriptor: | SULFATE ION, Uracil-DNA glycosylase | | Authors: | Earl, C, Bagneris, C, Cole, A.R, Barrett, T, Savva, R. | | Deposit date: | 2017-04-09 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structurally conserved motif in gamma-herpesvirus uracil-DNA glycosylases elicits duplex nucleotide-flipping.
Nucleic Acids Res., 46, 2018
|
|
5NN7
 
 | | KSHV uracil-DNA glycosylase, apo form | | Descriptor: | Uracil-DNA glycosylase | | Authors: | Earl, C, Bagneris, C, Cole, A.R, Barrett, T, Savva, R. | | Deposit date: | 2017-04-08 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structurally conserved motif in gamma-herpesvirus uracil-DNA glycosylases elicits duplex nucleotide-flipping.
Nucleic Acids Res., 46, 2018
|
|
5NNU
 
 | | KSHV uracil-DNA glycosylase, product complex with dsDNA exhibiting duplex nucleotide flipping | | Descriptor: | DNA, DNA containing an abasic site, Uracil-DNA glycosylase | | Authors: | Earl, C, Bagneris, C, Barrett, T, Savva, R. | | Deposit date: | 2017-04-10 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | A structurally conserved motif in gamma-herpesvirus uracil-DNA glycosylases elicits duplex nucleotide-flipping.
Nucleic Acids Res., 46, 2018
|
|
2OXM
 
 | | Crystal structure of a UNG2/modified DNA complex that represent a stabilized short-lived extrahelical state in ezymatic DNA base flipping | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*(4MF)P*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*AP*TP*CP*TP*T)-3'), Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Krosky, D.J, Stivers, J.T, Amzel, L.M. | | Deposit date: | 2007-02-20 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzymatic capture of an extrahelical thymine in the search for uracil in DNA.
Nature, 449, 2007
|
|
2OYT
 
 | | Crystal Structure of UNG2/DNA(TM) | | Descriptor: | DNA strand1, DNA strand2, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Krosky, D.J, Stivers, J.T, Amzel, L.M. | | Deposit date: | 2007-02-22 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymatic capture of an extrahelical thymine in the search for uracil in DNA.
Nature, 449, 2007
|
|
2RBA
 
 | | Structure of Human Thymine DNA Glycosylase Bound to Abasic and Undamaged DNA | | Descriptor: | DNA (5'-D(*DCP*DAP*DGP*DCP*DTP*DCP*DTP*DGP*DTP*DAP*DCP*DGP*DTP*DGP*DAP*DGP*DCP*DAP*DGP*DTP*DGP*DGP*DA)-3'), DNA (5'-D(*DCP*DCP*DAP*DCP*DTP*DGP*DCP*DTP*DCP*DAP*(3DR)P*DGP*DTP*DAP*DCP*DAP*DGP*DAP*DGP*DCP*DTP*DGP*DT)-3'), G/T mismatch-specific thymine DNA glycosylase | | Authors: | Maiti, A, Pozharski, E, Drohat, A.C. | | Deposit date: | 2007-09-18 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of human thymine DNA glycosylase bound to DNA elucidates sequence-specific mismatch recognition.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2HXM
 
 | | Complex of UNG2 and a small Molecule synthetic Inhibitor | | Descriptor: | 4-[(1E,7E)-8-(2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDIN-4-YL)-3,6-DIOXA-2,7-DIAZAOCTA-1,7-DIEN-1-YL]BENZOIC ACID, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Krosky, D.J, Ghung, S, Seiple, L, Amzel, L.M, Stivers, J.T. | | Deposit date: | 2006-08-03 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mimicking damaged DNA with a small molecule inhibitor of human UNG2.
Nucleic Acids Res., 34, 2006
|
|
2JHQ
 
 | | Crystal structure of Uracil DNA-glycosylase from Vibrio cholerae | | Descriptor: | CHLORIDE ION, URACIL DNA-GLYCOSYLASE | | Authors: | Raeder, I.L.U, Moe, E, Willassen, N.P, Smalas, A.O, Leiros, I. | | Deposit date: | 2007-02-23 | | Release date: | 2008-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Uracil-DNA N-Glycosylase (Ung) from Vibrio Cholerae. Mapping Temperature Adaptation Through Structural and Mutational Analysis.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2L3F
 
 | | Solution NMR Structure of a putative Uracil DNA glycosylase from Methanosarcina acetivorans, Northeast Structural Genomics Consortium Target MvR76 | | Descriptor: | Uncharacterized protein | | Authors: | Aramini, J.M, Hamilton, K, Ciccosanti, C.T, Wang, H, Lee, H.W, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-09-13 | | Release date: | 2010-10-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a putative Uracil DNA glycosylase from Methanosarcina acetivorans, Northeast Structural Genomics Consortium Target MvR76
To be Published
|
|
