1P2V
 
 | | H-RAS 166 in 60 % 1,6 hexanediol | | Descriptor: | HEXANE-1,6-DIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Buhrman, G.K, de Serrano, V, Mattos, C. | | Deposit date: | 2003-04-16 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Organic solvents order the dynamic switch II in Ras crystals
Structure, 11, 2003
|
|
1P2X
 
 | | CRYSTAL STRUCTURE OF THE CALPONIN-HOMOLOGY DOMAIN OF RNG2 FROM SCHIZOSACCHAROMYCES POMBE | | Descriptor: | BROMIDE ION, Ras GTPase-activating-like protein | | Authors: | Wang, C.-H, Balasubramanian, M.K, Dokland, T. | | Deposit date: | 2003-04-16 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure, crystal packing and molecular dynamics of the calponin-homology domain of Schizosaccharomyces pombe Rng2.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1P2Y
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME P450CAM IN COMPLEX WITH (S)-(-)-NICOTINE | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, Cytochrome P450-cam, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Strickler, M, Goldstein, B.M, Maxfield, K, Shireman, L, Kim, G, Matteson, D, Jones, J.P. | | Deposit date: | 2003-04-16 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic Studies on the Complex Behavior of Nicotine Binding to P450cam (CYP101)(dagger).
Biochemistry, 42, 2003
|
|
1P2Z
 
 | | Refinement of Adenovirus Type 2 Hexon with CNS | | Descriptor: | CITRIC ACID, Hexon protein | | Authors: | Rux, J.J, Kuser, P.R, Burnett, R.M. | | Deposit date: | 2003-04-16 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and phylogenetic analysis of adenovirus hexons by use of
high-resolution x-ray crystallographic, molecular modeling, and sequence-based
methods
J.VIROL., 77, 2003
|
|
1P30
 
 | | Refinement of Adenovirus Type 5 Hexon with CNS | | Descriptor: | Hexon protein | | Authors: | Rux, J.J, Kuser, P.R, Burnett, R.M. | | Deposit date: | 2003-04-16 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and phylogenetic analysis of adenovirus hexons by use of
high-resolution x-ray crystallographic, molecular modeling, and sequence-based
methods
J.VIROL., 77, 2003
|
|
1P31
 
 | | Crystal Structure of UDP-N-acetylmuramic acid:L-alanine Ligase (MurC) from Haemophilus influenzae | | Descriptor: | MAGNESIUM ION, UDP-N-acetylmuramate--alanine ligase, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Authors: | Mol, C.D, Brooun, A, Dougan, D.R, Hilgers, M.T, Tari, L.W, Wijnands, R.A, Knuth, M.W, McRee, D.E, Swanson, R.V. | | Deposit date: | 2003-04-16 | | Release date: | 2003-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Active Fully Assembled Substrate- and Product-Bound Complexes of UDP-N-Acetylmuramic Acid:L-Alanine Ligase (MurC) from Haemophilus influenzae.
J.Bacteriol., 185, 2003
|
|
1P33
 
 | | Pteridine reductase from Leishmania tarentolae complex with NADPH and MTX | | Descriptor: | METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase 1 | | Authors: | Zhao, H, Bray, T, Ouellette, M, Zhao, M, Ferre, R.A, Matthews, D, Whiteley, J.M, Varughese, K.I. | | Deposit date: | 2003-04-16 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure of pteridine reductase (PTR1) from Leishmania tarentolae.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1P36
 
 | | T4 LYOSZYME CORE REPACKING MUTANT I100V/TA | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-04-16 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Repacking the Core of T4 lysozyme by automated design
J.Mol.Biol., 332, 2003
|
|
1P37
 
 | | T4 LYSOZYME CORE REPACKING BACK-REVERTANT L102M/CORE10 | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-04-16 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Repacking the Core of T4 lysozyme by automated design
J.Mol.Biol., 332, 2003
|
|
1P39
 
 | |
1P3C
 
 | | Glutamyl endopeptidase from Bacillus intermedius | | Descriptor: | glutamyl-endopeptidase | | Authors: | Meijers, R, Blagova, E.V, Levdikov, V.M, Rudenskaya, G.N, Chestukhina, G.G, Akimkina, T.V, Kostrov, S.V, Lamzin, V.S, Kuranova, I.P. | | Deposit date: | 2003-04-17 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of glutamyl endopeptidase from Bacillus intermedius reveals a structural link between zymogen activation and charge compensation.
Biochemistry, 43, 2004
|
|
1P3D
 
 | | Crystal Structure of UDP-N-acetylmuramic acid:L-alanine ligase (MurC) in Complex with UMA and ANP. | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, UDP-N-acetylmuramate--alanine ligase, ... | | Authors: | Mol, C.D, Brooun, A, Dougan, D.R, Hilgers, M.T, Tari, L.W, Wijnands, R.A, Knuth, M.W, McRee, D.E, Swanson, R.V. | | Deposit date: | 2003-04-17 | | Release date: | 2003-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Active Fully Assembled Substrate- and Product-Bound Complexes of UDP-N-Acetylmuramic Acid:L-Alanine Ligase (MurC) from Haemophilus influenzae.
J.Bacteriol., 185, 2003
|
|
1P3E
 
 | | Structure of Glu endopeptidase in complex with MPD | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, glutamyl-endopeptidase | | Authors: | Meijers, R, Blagova, E.V, Levdikov, V.M, Rudenskaya, G.N, Chestukhina, G.G, Akimkina, T.V, Kostrov, S.V, Lamzin, V.S, Kuranova, I.P. | | Deposit date: | 2003-04-17 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The crystal structure of glutamyl endopeptidase from Bacillus intermedius reveals a structural link between zymogen activation and charge compensation.
Biochemistry, 43, 2004
|
|
1P3H
 
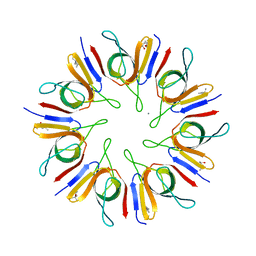 | | Crystal Structure of the Mycobacterium tuberculosis chaperonin 10 tetradecamer | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 10 kDa chaperonin, CALCIUM ION | | Authors: | Roberts, M.M, Coker, A.R, Fossati, G, Mascagni, P, Coates, A.R.M, Wood, S.P, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-17 | | Release date: | 2003-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mycobacterium tuberculosis chaperonin 10 heptamers self-associate through their biologically active loops
J.BACTERIOL., 185, 2003
|
|
1P3J
 
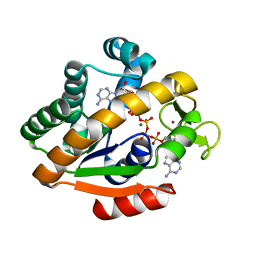 | | Adenylate Kinase from Bacillus subtilis | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bae, E, Phillips Jr, G.N. | | Deposit date: | 2003-04-17 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures and analysis of highly homologous psychrophilic, mesophilic, and thermophilic adenylate kinases.
J.Biol.Chem., 279, 2004
|
|
1P3N
 
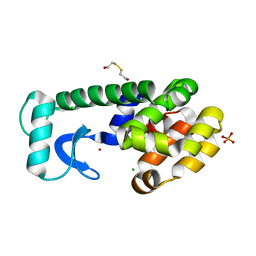 | | CORE REDESIGN BACK-REVERTANT I103V/CORE10 | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-04-17 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Repacking the Core of T4 lysozyme by automated design
J.Mol.Biol., 332, 2003
|
|
1P3Q
 
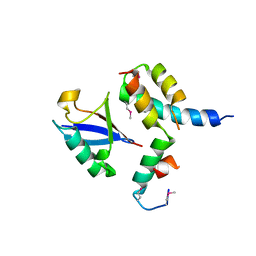 | | Mechanism of Ubiquitin Recognition by the CUE Domain of VPS9 | | Descriptor: | Ubiquitin, Vacuolar protein sorting-associated protein VPS9 | | Authors: | Prag, G, Misra, S, Jones, E.A, Ghirlando, R, Davies, B.A, Horazdovsky, B.F, Hurley, J.H. | | Deposit date: | 2003-04-18 | | Release date: | 2003-06-24 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Ubiquitin Recognition by the CUE Domain of Vps9p.
Cell(Cambridge,Mass.), 113, 2003
|
|
1P3W
 
 | |
1P3Y
 
 | | MrsD from Bacillus sp. HIL-Y85/54728 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MrsD protein | | Authors: | Blaesse, M, Kupke, T, Huber, R, Steinbacher, S. | | Deposit date: | 2003-04-19 | | Release date: | 2003-08-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structure of MrsD, an FAD-binding protein of the HFCD family.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1P42
 
 | | Crystal structure of Aquifex aeolicus LpxC Deacetylase (Zinc-Inhibited Form) | | Descriptor: | MYRISTIC ACID, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Whittington, D.A, Rusche, K.M, Shin, H, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2003-04-21 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of LpxC, a Zinc-Dependent Deacetylase Essential for Endotoxin Biosynthesis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1P44
 
 | | Targeting tuberculosis and malaria through inhibition of enoyl reductase: compound activity and structural data | | Descriptor: | 5-{[4-(9H-FLUOREN-9-YL)PIPERAZIN-1-YL]CARBONYL}-1H-INDOLE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kuo, M.R, Morbidoni, H.R, Alland, D, Sneddon, S.F, Gourlie, B.B, Staveski, M.M, Leonard, M, Gregory, J.S, Janjigian, A.D, Yee, C, Musser, J.M, Kreiswirth, B, Iwamoto, H, Perozzo, R, Jacobs Jr, W.R, Sacchettini, J.C, Fidock, D.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-21 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting tuberculosis and malaria through inhibition of Enoyl reductase: compound activity and structural data.
J.Biol.Chem., 278, 2003
|
|
1P45
 
 | | Targeting tuberculosis and malaria through inhibition of enoyl reductase: compound activity and structural data | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Kuo, M.R, Morbidoni, H.R, Alland, D, Sneddon, S.F, Gourlie, B.B, Staveski, M.M, Leonard, M, Gregory, J.S, Janjigian, A.D, Yee, C, Musser, J.M, Kreiswirth, B.N, Iwamoto, H, Perozzo, R, Jacobs Jr, W.R, Sacchettini, J.C, Fidock, D.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-21 | | Release date: | 2003-09-16 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Targeting tuberculosis and malaria through inhibition of Enoyl reductase: compound activity and structural data.
J.Biol.Chem., 278, 2003
|
|
1P46
 
 | | T4 lysozyme core repacking mutant M106I/TA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-04-21 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Repacking the Core of T4 lysozyme by automated design
J.Mol.Biol., 332, 2003
|
|
1P47
 
 | | Crystal Structure of tandem Zif268 molecules complexed to DNA | | Descriptor: | 5'-D(*CP*AP*CP*GP*CP*CP*CP*AP*CP*GP*CP*CP*GP*CP*CP*CP*AP*CP*GP*CP*CP*A)-3', 5'-D(*GP*TP*GP*GP*CP*GP*TP*GP*GP*GP*CP*GP*GP*CP*GP*TP*GP*GP*GP*CP*GP*T)-3', Early growth response protein 1, ... | | Authors: | Peisach, E, Pabo, C.O. | | Deposit date: | 2003-04-21 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Constraints for Zinc Finger Linker Design as Inferred from X-ray Crystal Structure of Tandem Zif268-DNA Complexes
J.Mol.Biol., 330, 2003
|
|
1P4A
 
 | | Crystal Structure of the PurR complexed with cPRPP | | Descriptor: | 1-ALPHA-PYROPHOSPHORYL-2-ALPHA,3-ALPHA-DIHYDROXY-4-BETA-CYCLOPENTANE-METHANOL-5-PHOSPHATE, Pur operon repressor | | Authors: | Bera, A.K, Zhu, J, Zalkin, H, Smith, J.L. | | Deposit date: | 2003-04-22 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Functional dissection of the Bacillus subtilis pur operator site.
J.Bacteriol., 185, 2003
|
|
