1M10
 
 | | Crystal structure of the complex of Glycoprotein Ib alpha and the von Willebrand Factor A1 Domain | | Descriptor: | Glycoprotein Ib alpha, von Willebrand Factor | | Authors: | Huizinga, E.G, Tsuji, S, Romijn, R.A.P, Schiphorst, M.E, de Groot, P.G, Sixma, J.J, Gros, P. | | Deposit date: | 2002-06-16 | | Release date: | 2002-08-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of glycoprotein Ibalpha and its complex with von Willebrand factor A1 domain.
Science, 297, 2002
|
|
1M13
 
 | | Crystal Structure of the Human Pregane X Receptor Ligand Binding Domain in Complex with Hyperforin, a Constituent of St. John's Wort | | Descriptor: | 4-HYDROXY-5-ISOBUTYRYL-6-METHYL-1,3,7-TRIS-(3-METHYL-BUT-2-ENYL)-6-(4-METHYL-PENT-3-ENYL)-BICYCLO[3.3.1]NON-3-ENE-2,9-DIONE, Orphan Nuclear Receptor PXR | | Authors: | Watkins, R.E, Maglich, J.M, Moore, L.B, Wisely, G.B, Noble, S.M, Davis-Searles, P.R, Lambert, M.H, Kliewer, S.A, Redinbo, M.R. | | Deposit date: | 2002-06-17 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.1 A Crystal Structure of Human PXR in Complex with the St. John's Wort Compound Hyperforin
Biochemistry, 42, 2003
|
|
1M14
 
 | |
1M15
 
 | | Transition state structure of arginine kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, MAGNESIUM ION, ... | | Authors: | Yousef, M.S, Fabiola, F, Gattis, J.L, Somasundaram, T, Chapman, M.S. | | Deposit date: | 2002-06-17 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Refinement of the arginine kinase transition-state analogue complex at 1.2 A resolution: mechanistic insights.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1M16
 
 | | Human Acidic Fibroblast Growth Factor. 141 Amino Acid Form with Amino Terminal His Tag and Leu 44 Replaced with Phe (L44F), Leu 73 Replaced with Val (L73V), Val 109 Replaced with Leu (V109L) and Cys 117 Replaced with Val (C117V). | | Descriptor: | FORMIC ACID, SULFATE ION, acidic fibroblast growth factor | | Authors: | Brych, S.R, Kim, J, Spielmann, G.L, Logan, T.M, Blaber, M. | | Deposit date: | 2002-06-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Accommodation of a highly symmetric core within a symmetric protein superfold
Protein Sci., 12, 2003
|
|
1M17
 
 | |
1M1B
 
 | | Crystal Structure of Phosphoenolpyruvate Mutase Complexed with Sulfopyruvate | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE PHOSPHOMUTASE, SULFOPYRUVATE | | Authors: | Liu, S, Lu, Z, Jia, Y, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2002-06-18 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dissociative phosphoryl transfer in PEP mutase catalysis: structure of the enzyme/sulfopyruvate complex and kinetic properties of mutants.
Biochemistry, 41, 2002
|
|
1M1C
 
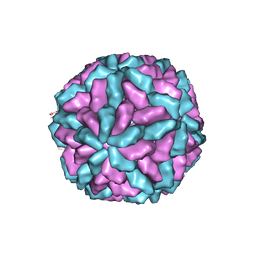 | | Structure of the L-A virus | | Descriptor: | Major coat protein | | Authors: | Naitow, H, Tang, J, Canady, M, Wickner, R.B, Johnson, J.E. | | Deposit date: | 2002-06-18 | | Release date: | 2002-10-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | L-A virus at 3.4 A resolution reveals particle architecture and mRNA decapping mechanism.
Nat.Struct.Biol., 9, 2002
|
|
1M1E
 
 | | Beta-catenin armadillo repeat domain bound to ICAT | | Descriptor: | Beta-catenin, ICAT | | Authors: | Daniels, D.L, Weis, W.I. | | Deposit date: | 2002-06-18 | | Release date: | 2002-10-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ICAT inhibits Beta-catenin binding to Tcf/Lef-family transcription factors and the general coactivator p300 using independent structural modules.
Mol.Cell, 10, 2002
|
|
1M1F
 
 | | Kid toxin protein from E.coli plasmid R1 | | Descriptor: | Kid toxin protein, PHOSPHATE ION | | Authors: | Hargreaves, D, Santos-Sierra, S, Giraldo, R, Sabariegos-Jareno, R, de la Cueva-Mendez, G, Boelens, R, Diaz-Orejas, R, Rafferty, J.B. | | Deposit date: | 2002-06-19 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Functional Analysis of the Kid toxin protein from E.coli plasmid R1
Structure, 10, 2002
|
|
1M1K
 
 | | Co-crystal structure of azithromycin bound to the 50S ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, AZITHROMYCIN, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2002-06-19 | | Release date: | 2002-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
1M1L
 
 | | Human Suppressor of Fused (N-terminal domain) | | Descriptor: | Suppressor of Fused | | Authors: | Merchant, M, Vajdos, F.F, Ultsch, M, Maun, H.R, Wendt, U, Cannon, J, Lazarus, R.A, de Vos, A.M, de Sauvage, F.J. | | Deposit date: | 2002-06-19 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Suppressor of fused regulates Gli activity through a dual binding mechanism
Mol.Cell.Biol., 24, 2004
|
|
1M1M
 
 | |
1M1O
 
 | | Crystal structure of biosynthetic thiolase, C89A mutant, complexed with acetoacetyl-CoA | | Descriptor: | ACETOACETYL-COENZYME A, Acetyl-CoA acetyltransferase, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-06-20 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
1M1P
 
 | | P21 crystal structure of the tetraheme cytochrome c3 from Shewanella oneidensis MR1 | | Descriptor: | HEME C, SULFATE ION, Small tetraheme cytochrome c | | Authors: | Leys, D, Meyer, T.E, Tsapin, A.I, Nealson, K.H, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2002-06-20 | | Release date: | 2002-08-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures at atomic resolution reveal the novel concept of 'electron-harvesting' as a role for the small tetraheme cytochrome c
J.Biol.Chem., 277, 2002
|
|
1M1Q
 
 | | P222 oxidized structure of the tetraheme cytochrome c from Shewanella oneidensis MR1 | | Descriptor: | HEME C, SULFATE ION, small tetraheme cytochrome c | | Authors: | Leys, D, Meyer, T.E, Tsapin, A.I, Nealson, K.H, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2002-06-20 | | Release date: | 2002-08-14 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal structures at atomic resolution reveal the novel concept of 'electron-harvesting' as a role for the small tetraheme cytochrome c
J.Biol.Chem., 277, 2002
|
|
1M1R
 
 | | Reduced p222 crystal structure of the tetraheme cytochrome c of Shewanella oneidensis MR1 | | Descriptor: | HEME C, SMALL tetraheme cytochrome c, SULFATE ION | | Authors: | Leys, D, Meyer, T.E, Tsapin, A.I, Nealson, K.H, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2002-06-20 | | Release date: | 2002-08-14 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structures at atomic resolution reveal the novel concept of 'electron-harvesting' as a role for the small tetraheme cytochrome c
J.Biol.Chem., 277, 2002
|
|
1M1S
 
 | | Structure of WR4, a C.elegans MSP family member | | Descriptor: | WR4 | | Authors: | Karpowich, N, Smith, P, Shen, J, Hunt, J, Montelione, G, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-20 | | Release date: | 2003-07-29 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a C.elegans MSP family member
To be Published
|
|
1M1T
 
 | | Biosynthetic thiolase, Q64A mutant | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-06-20 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
1M1Z
 
 | |
1M20
 
 | | Crystal Structure of F35Y Mutant of Trypsin-solubilized Fragment of Cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yao, P, Wu, J, Wang, Y.-H, Sun, B.-Y, Xia, Z.-X, Huang, Z.-X. | | Deposit date: | 2002-06-20 | | Release date: | 2002-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallography, CD and kinetic studies revealed the essence of the abnormal behaviors of the cytochrome b5 Phe35-->Tyr mutant.
Eur.J.Biochem., 269, 2002
|
|
1M21
 
 | | Crystal structure analysis of the peptide amidase PAM in complex with the competitive inhibitor chymostatin | | Descriptor: | CHYMOSTATIN, Peptide Amidase | | Authors: | Labahn, J, Neumann, S, Buldt, G, Kula, M.-R, Granzin, J. | | Deposit date: | 2002-06-21 | | Release date: | 2002-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An alternative mechanism for amidase signature enzymes
J.MOL.BIOL., 322, 2002
|
|
1M22
 
 | | X-ray structure of native peptide amidase from Stenotrophomonas maltophilia at 1.4 A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, peptide amidase | | Authors: | Labahn, J, Neumann, S, Buldt, G, Kula, M.-R, Granzin, J. | | Deposit date: | 2002-06-21 | | Release date: | 2002-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An alternative mechanism for amidase signature enzymes
J.MOL.BIOL., 322, 2002
|
|
1M24
 
 | |
1M26
 
 | | Crystal structure of jacalin-T-antigen complex | | Descriptor: | Jacalin, alpha chain, beta chain, ... | | Authors: | Jeyaprakash, A.A, Rani, P.G, Reddy, G.B, Banumathi, S, Betzel, C, Surolia, A, Vijayan, M. | | Deposit date: | 2002-06-21 | | Release date: | 2002-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of the jacalin-T-antigen complex and a
comparative study of lectin-T-antigen complexs
J.Mol.Biol., 321, 2002
|
|
