4N8Y
 
 | | Crystal structure of a trap periplasmic solute binding protein from bradyrhizobium sp. btai1 b (bbta_0128), target EFI-510056 (bbta_0128), complex with alpha/beta-d-galacturonate | | Descriptor: | Putative TRAP-type C4-dicarboxylate transport system, binding periplasmic protein (DctP subunit), alpha-D-galactopyranuronic acid, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-18 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N8Z
 
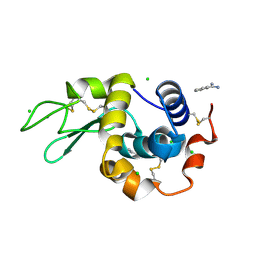 | | In situ lysozyme crystallized on a MiTeGen micromesh with benzamidine ligand | | Descriptor: | BENZAMIDINE, CHLORIDE ION, Lysozyme C, ... | | Authors: | Yin, X, Scalia, A, Leroy, L, Cuttitta, C.M, Polizzo, G.M, Ericson, D.L, Roessler, C.G, Campos, O, Agarwal, R, Allaire, M, Orville, A.M, Jackimowicz, R, Ma, M.Y, Sweet, R.M, Soares, A.S. | | Deposit date: | 2013-10-18 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Hitting the target: fragment screening with acoustic in situ co-crystallization of proteins plus fragment libraries on pin-mounted data-collection micromeshes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N90
 
 | |
4N91
 
 | | Crystal structure of a trap periplasmic solute binding protein from anaerococcus prevotii dsm 20548 (Apre_1383), target EFI-510023, with bound alpha/beta d-glucuronate | | Descriptor: | CHLORIDE ION, TETRAETHYLENE GLYCOL, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-18 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N92
 
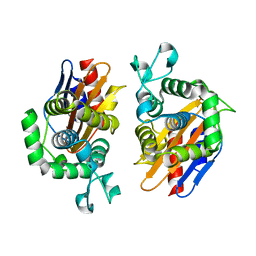 | | Crystal structure of beta-lactamse PenP_E166S | | Descriptor: | Beta-lactamase | | Authors: | Pan, X, Wong, W, Zhao, Y. | | Deposit date: | 2013-10-19 | | Release date: | 2014-09-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Perturbing the General Base Residue Glu166 in the Active Site of Class A beta-Lactamase Leads to Enhanced Carbapenem Binding and Acylation
Biochemistry, 53, 2014
|
|
4N93
 
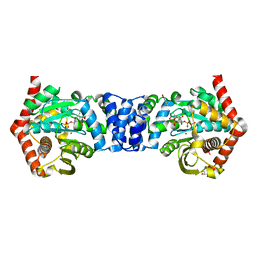 | | Alternative substrates of Mycobacterium tuberculosis anthranilate phosphoribosyl transferase | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-amino-6-methylbenzoic acid, Anthranilate phosphoribosyltransferase, ... | | Authors: | Castell, A, Short, F.L, Lott, J.S. | | Deposit date: | 2013-10-19 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Alternative substrates reveal catalytic cycle and key binding events in the reaction catalysed by anthranilate phosphoribosyltransferase from Mycobacterium tuberculosis.
Biochem.J., 461, 2014
|
|
4N94
 
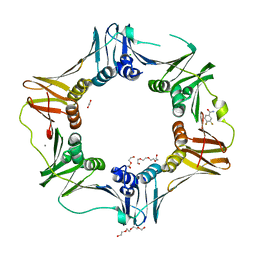 | | E. coli sliding clamp in complex with 3,4-difluorobenzamide | | Descriptor: | 1,2-ETHANEDIOL, 3,4-difluorobenzamide, CALCIUM ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4N95
 
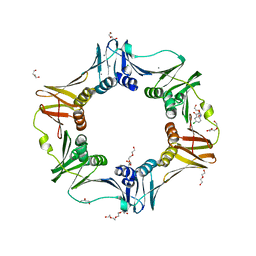 | | E. coli sliding clamp in complex with 5-chloroindoline-2,3-dione | | Descriptor: | 5-chloro-1H-indole-2,3-dione, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4N96
 
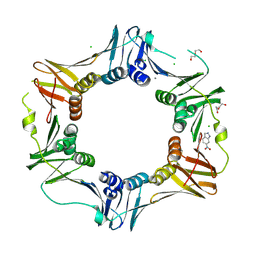 | | E. coli sliding clamp in complex with 6-nitroindazole | | Descriptor: | 6-NITROINDAZOLE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4N97
 
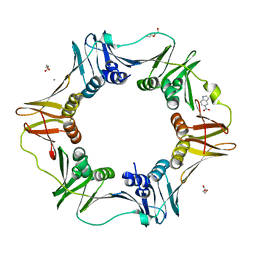 | | E. coli sliding clamp in complex with 5-nitroindole | | Descriptor: | 5-nitro-1H-indole, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4N98
 
 | |
4N99
 
 | | E. coli sliding clamp in complex with 6-chloro-2,3,4,9-tetrahydro-1H-carbazole-7-carboxylic acid | | Descriptor: | 6-chloro-2,3,4,9-tetrahydro-1H-carbazole-7-carboxylic acid, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4N9A
 
 | | E. coli sliding clamp in complex with (R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxylic acid | | Descriptor: | (1R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4N9B
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1-methyl-N-(pyridin-3-yl)-1H-pyrazole-5-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhai, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.859 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N9C
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 5-nitro-1H-benzimidazole, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N9D
 
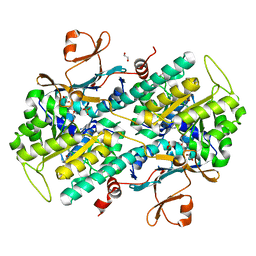 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, 4-({[(4-tert-butylphenyl)sulfonyl]amino}methyl)-N-(pyridin-3-yl)benzamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N9E
 
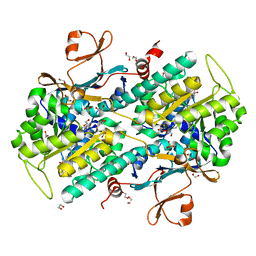 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1-benzoylpiperidin-4-yl)methyl]-N-(pyridin-3-yl)-1H-benzimidazole-5-carboxamide, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | Deposit date: | 2013-10-20 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N9F
 
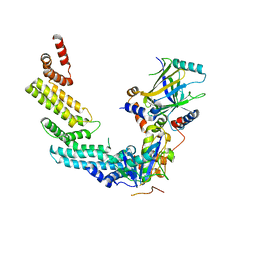 | | Crystal structure of the Vif-CBFbeta-CUL5-ElOB-ElOC pentameric complex | | Descriptor: | Core-binding factor subunit beta, Cullin-5, Transcription elongation factor B polypeptide 1, ... | | Authors: | Guo, Y.Y, Dong, L.Y, Huang, Z.W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for hijacking CBF-b and CUL5 E3 ligase complex by HIV-1 Vif
Nature, 505, 2014
|
|
4N9G
 
 | |
4N9H
 
 | | Crystal structure of Transcription regulation Protein CRP | | Descriptor: | Catabolite gene activator | | Authors: | Lee, B.J, Seok, S.H, Im, H, Yoon, H.J. | | Deposit date: | 2013-10-21 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of inactive CRP species reveal the atomic details of the allosteric transition that discriminates cyclic nucleotide second messengers.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N9I
 
 | | Crystal Structure of Transcription regulation protein CRP complexed with cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Catabolite gene activator | | Authors: | Lee, B.-J, Seok, S.-H, Im, H, Yoon, H.-J. | | Deposit date: | 2013-10-21 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structures of inactive CRP species reveal the atomic details of the allosteric transition that discriminates cyclic nucleotide second messengers.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N9J
 
 | | Crystal structure of the cryptic polo box domain of human Plk4 | | Descriptor: | CHLORIDE ION, SULFATE ION, Serine/threonine-protein kinase PLK4 | | Authors: | Ku, B, Kim, J.H, Lee, K.S, Kim, S.J. | | Deposit date: | 2013-10-21 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4N9K
 
 | | crystal structure of beta-lactamse PenP_E166S in complex with cephaloridine | | Descriptor: | 5-METHYL-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Pan, X, Wong, W, Zhao, Y. | | Deposit date: | 2013-10-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Perturbing the General Base Residue Glu166 in the Active Site of Class A beta-Lactamase Leads to Enhanced Carbapenem Binding and Acylation
Biochemistry, 53, 2014
|
|
4N9L
 
 | | crystal structure of beta-lactamse PenP_E166S in complex with meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase | | Authors: | Pan, X, Wong, W, Zhao, Y. | | Deposit date: | 2013-10-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Perturbing the General Base Residue Glu166 in the Active Site of Class A beta-Lactamase Leads to Enhanced Carbapenem Binding and Acylation
Biochemistry, 53, 2014
|
|
4N9M
 
 | | Joint neutron/x-ray structure of urate oxidase in complex with 8-hydroxyxanthine | | Descriptor: | 8-hydroxy-3,9-dihydro-1H-purine-2,6-dione, CHLORIDE ION, SODIUM ION, ... | | Authors: | Oksanen, E, Blakeley, M.P, Budayova-Spano, M. | | Deposit date: | 2013-10-21 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-20 | | Method: | NEUTRON DIFFRACTION (2.023 Å), X-RAY DIFFRACTION | | Cite: | The neutron structure of urate oxidase resolves a long-standing mechanistic conundrum and reveals unexpected changes in protonation.
Plos One, 9, 2014
|
|
