4N86
 
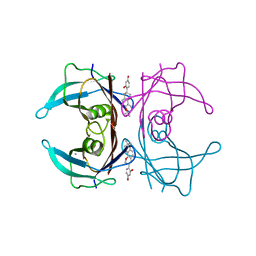 | | Crystal structure of human transthyretin complexed with glabridin | | Descriptor: | 4-[(3R)-8,8-dimethyl-3,4-dihydro-2H,8H-pyrano[2,3-f]chromen-3-yl]benzene-1,3-diol, CALCIUM ION, Transthyretin | | Authors: | Yokoyama, T, Kosaka, Y, Mizuguchi, M. | | Deposit date: | 2013-10-17 | | Release date: | 2014-02-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structures of human transthyretin complexed with glabridin
J.Med.Chem., 57, 2014
|
|
4N87
 
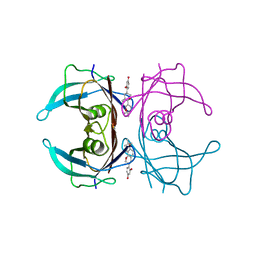 | | Crystal structure of V30M mutant human transthyretin complexed with glabridin | | Descriptor: | 4-[(3R)-8,8-dimethyl-3,4-dihydro-2H,8H-pyrano[2,3-f]chromen-3-yl]benzene-1,3-diol, Transthyretin | | Authors: | Yokoyama, T, Kosaka, Y, Mizuguchi, M. | | Deposit date: | 2013-10-17 | | Release date: | 2014-02-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Crystal structures of human transthyretin complexed with glabridin
J.Med.Chem., 57, 2014
|
|
4N88
 
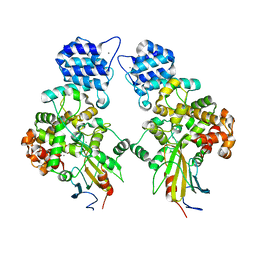 | | Crystal structure of Tse3-Tsi3 complex with calcium ion | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | Shang, G.J. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
4N8C
 
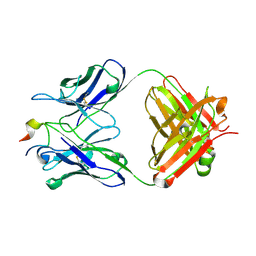 | | Three-dimensional structure of the extracellular domain of Matrix protein 2 of influenza A virus | | Descriptor: | Extracellular domain of influenza Matrix protein 2, Heavy chain of monoclonal antibody, Light chain of monoclonal antibody | | Authors: | Cho, K.J, Seok, J.H, Kim, S, Roose, K, Schepens, B, Fiers, W, Saelens, X, Kim, K.H. | | Deposit date: | 2013-10-17 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the extracellular domain of matrix protein 2 of influenza A virus in complex with a protective monoclonal antibody
J.Virol., 89, 2015
|
|
4N8D
 
 | | DPP4 complexed with syn-7aa | | Descriptor: | 1-(cis-1-phenyl-4-{[(2E)-3-phenylprop-2-en-1-yl]oxy}cyclohexyl)methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Ostermann, N, Zink, F, Kroemer, M. | | Deposit date: | 2013-10-17 | | Release date: | 2014-02-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of C-(1-aryl-cyclohexyl)-methylamines as selective, orally available inhibitors of dipeptidyl peptidase IV.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N8E
 
 | | DPP4 complexed with compound 12a | | Descriptor: | 1-[cis-4-(aminomethyl)-4-(3-chlorophenyl)cyclohexyl]piperidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Ostermann, N, Zink, F, Kroemer, M. | | Deposit date: | 2013-10-17 | | Release date: | 2014-02-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of C-(1-aryl-cyclohexyl)-methylamines as selective, orally available inhibitors of dipeptidyl peptidase IV.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N8F
 
 | | CcmL from Thermosynechococcus elongatus BP-1 | | Descriptor: | Carbon dioxide concentrating mechanism protein, MAGNESIUM ION, SULFATE ION | | Authors: | Kimber, M.S, Demers, R.J. | | Deposit date: | 2013-10-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interactions and structural variability of beta-carboxysomal shell protein CcmL.
Photosynth.Res., 121, 2014
|
|
4N8G
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0660), Target EFI-501075, with bound D-alanine-D-alanine | | Descriptor: | CHLORIDE ION, D-ALANINE, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N8H
 
 | | E61V mutant, RipA structure | | Descriptor: | 4-hydroxybutyrate coenzyme A transferase | | Authors: | Torres, R, Goulding, C.W. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural snapshots along the reaction pathway of Yersinia pestis RipA, a putative butyryl-CoA transferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N8I
 
 | | M31G mutant, RipA structure | | Descriptor: | 4-hydroxybutyrate coenzyme A transferase, ACETATE ION, COENZYME A | | Authors: | Torres, R, Goulding, C.W. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Structural snapshots along the reaction pathway of Yersinia pestis RipA, a putative butyryl-CoA transferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N8J
 
 | | F60M mutant, RipA structure | | Descriptor: | ACETATE ION, COENZYME A, PHOSPHATE ION, ... | | Authors: | Torres, R, Goulding, C.W. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural snapshots along the reaction pathway of Yersinia pestis RipA, a putative butyryl-CoA transferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N8K
 
 | | E249A mutant, RipA structure | | Descriptor: | Putative 4-hydroxybutyrate coenzyme A transferase | | Authors: | Torres, R, Goulding, C.W. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural snapshots along the reaction pathway of Yersinia pestis RipA, a putative butyryl-CoA transferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N8L
 
 | | E249D mutant, RipA structure | | Descriptor: | Putative 4-hydroxybutyrate coenzyme A transferase | | Authors: | Torres, R, Goulding, C.W. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.817 Å) | | Cite: | Structural snapshots along the reaction pathway of Yersinia pestis RipA, a putative butyryl-CoA transferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N8M
 
 | | Structural polymorphism in the N-terminal oligomerization domain of NPM1 | | Descriptor: | COBALT (II) ION, Nucleophosmin | | Authors: | Mitrea, D, Royappa, G, Buljan, M, Yun, M, Pytel, N, Satumba, J, Nourse, A, Park, C, Babu, M.M, White, S.W, Kriwacki, R.W. | | Deposit date: | 2013-10-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural polymorphism in the N-terminal oligomerization domain of NPM1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4N8N
 
 | | Crystal structure of Mycobacterial FtsX extracellular domain | | Descriptor: | Cell division protein FtsX, POTASSIUM ION | | Authors: | Mavrici, D. | | Deposit date: | 2013-10-17 | | Release date: | 2014-05-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.874 Å) | | Cite: | Mycobacterium tuberculosis FtsX extracellular domain activates the peptidoglycan hydrolase, RipC.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4N8O
 
 | |
4N8P
 
 | | Crystal structure of a strand swapped CTLA-4 from Duckbill Platypus [PSI-NYSGRC-012711] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Uncharacterized protein | | Authors: | Kumar, P.R, Banu, R, Bhosle, R, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Glenn, A.S, Hillerich, B, Khafizov, K, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Crystal structure of a strand swapped CTLA-4 from Duckbill Platypus [PSI-NYSGRC-012711]
to be published
|
|
4N8Q
 
 | | Alternative substrates of Mycobacterium tuberculosis anthranilate phosphoribosyl transferase | | Descriptor: | 2-amino-4-fluorobenzoic acid, Anthranilate phosphoribosyltransferase | | Authors: | Castell, A, Cookson, T.V.M, Bulloch, E, Evans, G.L, Baker, E.N, Lott, J.S, Parker, E.J. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Alternative substrates reveal catalytic cycle and key binding events in the reaction catalysed by anthranilate phosphoribosyltransferase from Mycobacterium tuberculosis.
Biochem.J., 461, 2014
|
|
4N8R
 
 | | Crystal structure of RXRa LBD complexed with a synthetic modulator K-8008 | | Descriptor: | 5-(2-{(1Z)-2-methyl-1-[4-(propan-2-yl)benzylidene]-1H-inden-3-yl}ethyl)-1H-tetrazole, Retinoic acid receptor RXR-alpha | | Authors: | Aleshin, A.E, Su, Y, Zhang, X, Liddington, R.C. | | Deposit date: | 2013-10-17 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Sulindac-Derived RXR alpha Modulators Inhibit Cancer Cell Growth by Binding to a Novel Site.
Chem.Biol., 21, 2014
|
|
4N8S
 
 | | Crystal Structure of the ternary complex of lipase from Thermomyces lanuginosa with Ethylacetoacetate and P-nitrobenzaldehyde at 2.3 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitrobenzaldehyde, GLYCEROL, ... | | Authors: | Kumar, M, Mukherjee, J, Gupta, M.N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-10-18 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the ternary complex of lipase from Thermomyces lanuginosa with Ethylacetoacetate and P-nitrobenzaldehyde at 2.3 A resolution
To be Published
|
|
4N8T
 
 | | Human hemoglobin nitric oxide adduct | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, NITRIC OXIDE, ... | | Authors: | Yi, J, Richter-Addo, G.B. | | Deposit date: | 2013-10-18 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic characterization of the nitric oxide derivative of R-state human hemoglobin.
Nitric Oxide, 39, 2014
|
|
4N8U
 
 | | Two-Domain Laccase from Streptomyces viridochromogenes at 2.4 A resolution AC629 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Gabdulkhakov, A, Tischenko, S, Yurevich, L, Lisov, A, Leontievsky, A. | | Deposit date: | 2013-10-18 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two-Domain Laccase from Streptomyces viridochromogenes at 2.4 A resolution AC629
To be Published
|
|
4N8V
 
 | |
4N8W
 
 | | cathepsin K - chondroitin sulfate complex | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, Cathepsin K | | Authors: | Aguda, A.H, Nguyen, N.T, Bromme, D, Brayer, G.D. | | Deposit date: | 2013-10-18 | | Release date: | 2014-11-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis of collagen fiber degradation by cathepsin K.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4N8X
 
 | | The structure of Nostoc sp. PCC 7120 CcmL | | Descriptor: | Carbon dioxide concentrating mechanism protein, SULFATE ION | | Authors: | Keeling, T.J, Kimber, M.S. | | Deposit date: | 2013-10-18 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Interactions and structural variability of beta-carboxysomal shell protein CcmL.
Photosynth.Res., 121, 2014
|
|
