2N89
 
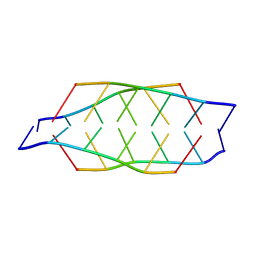 | | Tetrameric i-motif structure of dT-dC-dC-CFL-CFL-dC at acidic pH | | Descriptor: | DNA (5'-D(*TP*CP*CP*(CFL)P*(CFL)P*C)-3') | | Authors: | Abou-Assi, H, Harkness, R.W, Martin-Pintado, N, Wilds, C.J, Campos-Olivas, R, Mittermaier, A.K, Gonzalez, C, Damha, M.J. | | Deposit date: | 2015-10-09 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Stabilization of i-motif structures by 2'-beta-fluorination of DNA.
Nucleic Acids Res., 44, 2016
|
|
2N8A
 
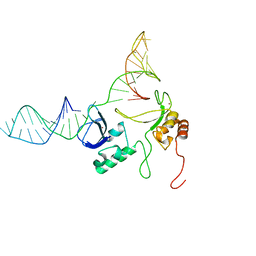 | | 1H, 13C and 15N chemical shift assignments and solution structure for PARP-1 F1F2 domains in complex with a DNA single-strand break | | Descriptor: | DNA (45-MER), Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Yang, J, Wu, W. | | Deposit date: | 2015-10-08 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Detection and Signaling of DNA Single-Strand Breaks by Human PARP-1.
Mol.Cell, 60, 2015
|
|
2N8B
 
 | |
2N8C
 
 | |
2N8D
 
 | |
2N8E
 
 | |
2N8F
 
 | |
2N8G
 
 | | NMR Structure of the homeodomain transcription factor Gbx1[E23R,R58E] from Homo sapiens | | Descriptor: | Homeobox protein GBX-1 | | Authors: | Proudfoot, A.K, Serrano, P, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG), Partnership for Stem Cell Biology (STEMCELL) | | Deposit date: | 2015-10-15 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the homeodomain transcription factor Gbx1[E23R,R58E] from Homo sapiens
To be Published
|
|
2N8H
 
 | |
2N8I
 
 | | Solution NMR Structure of Designed Protein DA05, Northeast Structural Genomics Consortium (NESG) Target OR626 | | Descriptor: | Designed Protein DA05 | | Authors: | Xu, X, Eletsky, A, Federizon, J.F, Jacobs, T.M, Kuhlman, B, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-10-15 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of structurally distinct proteins using strategies inspired by evolution.
Science, 352, 2016
|
|
2N8J
 
 | |
2N8K
 
 | | Chemical Shift Assignments and Structure Determination for spider toxin, U33-theraphotoxin-Cg1c | | Descriptor: | U33-theraphotoxin-Cg1b | | Authors: | Chin, Y.K.-Y, Pineda, S.S, Mobli, M, King, G.F. | | Deposit date: | 2015-10-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural venomics reveals evolution of a complex venom by duplication and diversification of an ancient peptide-encoding gene.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2N8L
 
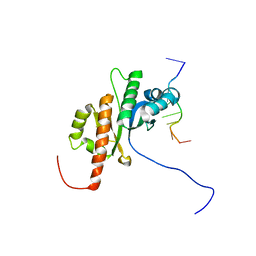 | |
2N8M
 
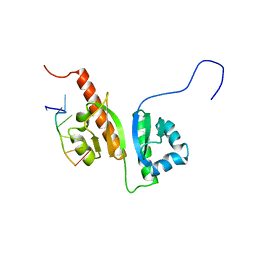 | |
2N8N
 
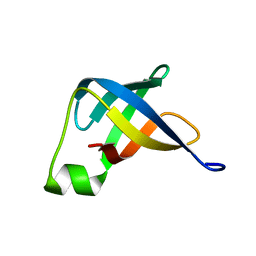 | | Solution structure of translation initiation factor | | Descriptor: | Translation initiation factor IF-1 | | Authors: | Kim, D, Lee, K, Lee, B. | | Deposit date: | 2015-10-21 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics study of translation initiation factor 1 from Staphylococcus aureus suggests its RNA binding mode
Biochim.Biophys.Acta, 1865, 2017
|
|
2N8O
 
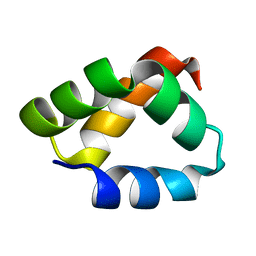 | | NMR Solution Structure of Aureocin A53 | | Descriptor: | Bacteriocin aureocin A53 | | Authors: | Acedo, J.Z, van Belkum, M.J, Lohans, C.T, Towle, K.M, Miskolzie, M, Vederas, J.C. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structures of Lacticin Q and Aureocin A53 Reveal a Structural Motif Conserved among Leaderless Bacteriocins with Broad-Spectrum Activity.
Biochemistry, 55, 2016
|
|
2N8P
 
 | | Solution Structure of Lacticin Q | | Descriptor: | Lacticin Q | | Authors: | Acedo, J.Z, van Belkum, M.J, Lohans, C.T, Towle, K.M, Miskolzie, M, Vederas, J.C. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structures of Lacticin Q and Aureocin A53 Reveal a Structural Motif Conserved among Leaderless Bacteriocins with Broad-Spectrum Activity.
Biochemistry, 55, 2016
|
|
2N8Q
 
 | |
2N8R
 
 | |
2N8S
 
 | |
2N8T
 
 | |
2N8U
 
 | |
2N8V
 
 | | An NMR/SAXS structure of the PKI domain of the honeybee dicistrovirus, Israeli acute paralysis virus (IAPV) IRES | | Descriptor: | RNA (70-MER) | | Authors: | Au, H.H, Cornilescu, G, Mouzakis, K.D, Burke, J.E, Ren, Q, Lee, S, Butcher, S.E, Jan, E. | | Deposit date: | 2015-10-27 | | Release date: | 2015-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Global shape mimicry of tRNA within a viral internal ribosome entry site mediates translational reading frame selection.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2N8W
 
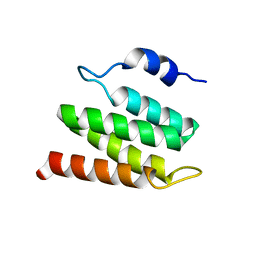 | | Solution NMR Structure of Designed Protein DA05R1, Northeast Structural Genomics Consortium (NESG) Target OR690 | | Descriptor: | Designed Protein DA05R1 | | Authors: | Eletsky, A, Federizon, J.F, Xu, X, Pulavarti, S, Jacobs, T.M, Kuhlman, B, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-10-27 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of structurally distinct proteins using strategies inspired by evolution.
Science, 352, 2016
|
|
2N8X
 
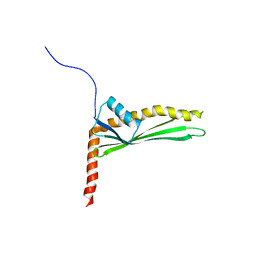 | | Solution structure of LptE from Pseudomonas Aerigunosa | | Descriptor: | LPS-assembly lipoprotein LptE | | Authors: | Moehle, K, Kocherla, H, Jurt, S, Robinson, J, Zerbe, O, Zerbe, K, Bacsa, B. | | Deposit date: | 2015-10-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of LptE from Pseudomonas aeruginosa.
Biochemistry, 55, 2016
|
|
