7CJV
 
 | | Solution structure of monomeric superoxide dismutase 1 with an additional mutation H46W in a dilute environment | | Descriptor: | Monomeric Human Cu,Zn Superoxide dismutase | | Authors: | Iwakawa, N, Morimoto, D, Walinda, E, Danielsson, J, Shirakawa, M, Sugase, K. | | Deposit date: | 2020-07-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Transient Diffusive Interactions with a Protein Crowder Affect Aggregation Processes of Superoxide Dismutase 1 beta-Barrel.
J.Phys.Chem.B, 125, 2021
|
|
7CJW
 
 | | Solution structure of monomeric superoxide dismutase 1 with an additional mutation H46W in a crowded environment | | Descriptor: | Monomeric Human Cu,Zn Superoxide dismutase | | Authors: | Iwakawa, N, Morimoto, D, Walinda, E, Danielsson, J, Shirakawa, M, Sugase, K. | | Deposit date: | 2020-07-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Transient Diffusive Interactions with a Protein Crowder Affect Aggregation Processes of Superoxide Dismutase 1 beta-Barrel.
J.Phys.Chem.B, 125, 2021
|
|
7CK5
 
 | | Solution structure of 28 amino acid polypeptide (354-381) in Plantago asiatica mosaic virus replicase bound to SDS micelle | | Descriptor: | PlAMV replicase peptide from RNA-dependent RNA polymerase | | Authors: | Komatsu, K, Sasaki, N, Yoshida, T, Suzuki, K, Masujima, Y, Hashimoto, M, Watanabe, S, Tochio, N, Kigawa, T, Yamaji, Y, Oshima, K, Namba, S, Nelson, R, Arie, T. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification of a Proline-Kinked Amphipathic alpha-Helix Downstream from the Methyltransferase Domain of a Potexvirus Replicase and Its Role in Virus Replication and Perinuclear Complex Formation.
J.Virol., 95, 2021
|
|
7CKD
 
 | |
7CKE
 
 | |
7CLS
 
 | |
7CLV
 
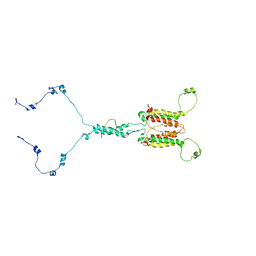 | | Solution structure of mitochondrial Tim23 channel in complex with a signaling peptide | | Descriptor: | COX4 isoform 1, TIM23 isoform 1 | | Authors: | Zhou, S, Ruan, M.S, Li, Y.Y, Yang, J, Richter, C, Schwalbe, H, Shen, B, Wang, J.F. | | Deposit date: | 2020-07-22 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the voltage-gated Tim23 channel in complex with a mitochondrial presequence peptide.
Cell Res., 31, 2021
|
|
7CNF
 
 | | Solution strcture of HsTFIIS LW domain | | Descriptor: | Transcription elongation factor A protein 1 | | Authors: | Gao, J, Liao, S. | | Deposit date: | 2020-07-31 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution strcture of HsTFIIS LW domain
To Be Published
|
|
7CPS
 
 | |
7CQ1
 
 | |
7CSK
 
 | |
7CSQ
 
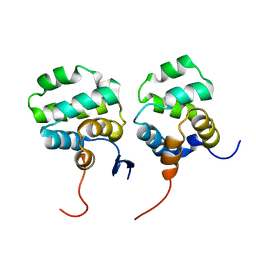 | | Solution structure of the complex between p75NTR-DD and TRADD-DD | | Descriptor: | Tumor necrosis factor receptor superfamily member 16, Tumor necrosis factor receptor type 1-associated DEATH domain protein | | Authors: | Lin, Z, Zhang, N. | | Deposit date: | 2020-08-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of NF-kappa B signaling by the p75 neurotrophin receptor interaction with adaptor protein TRADD through their respective death domains.
J.Biol.Chem., 297, 2021
|
|
7CSS
 
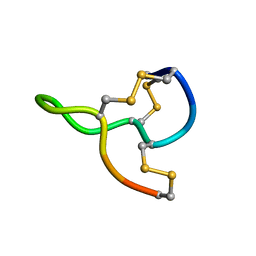 | |
7CU6
 
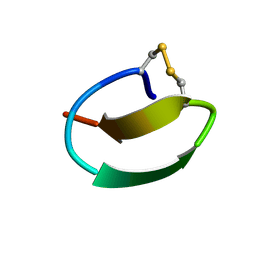 | | lasso peptide C24 mutant - A11V2C | | Descriptor: | lasso peptide C24_A11V2C | | Authors: | Ma, M, Liu, X.H. | | Deposit date: | 2020-08-21 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7CUK
 
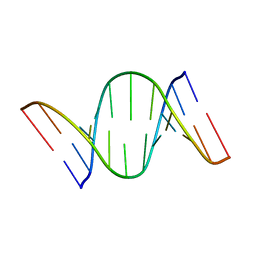 | |
7CV3
 
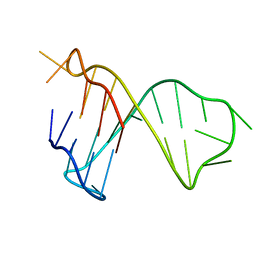 | | Quadruplex-duplex hybrid structure in the PIM1 gene, Form 1 | | Descriptor: | PIM1 promoter, Form 1 | | Authors: | Winnerdy, F.R, Tan, D.J.Y, Lim, K.W, Phan, A.T. | | Deposit date: | 2020-08-25 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Coexistence of two quadruplex-duplex hybrids in the PIM1 gene.
Nucleic Acids Res., 48, 2020
|
|
7CV4
 
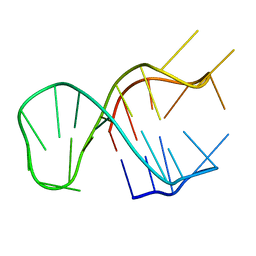 | | Quadruplex-duplex hybrid structure in the PIM1 gene, Form 2 | | Descriptor: | DNA (26-MER) | | Authors: | Winnerdy, F.R, Tan, D.T.J, Lim, K.W, Phan, A.T. | | Deposit date: | 2020-08-25 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Coexistence of two quadruplex-duplex hybrids in the PIM1 gene.
Nucleic Acids Res., 48, 2020
|
|
7CWH
 
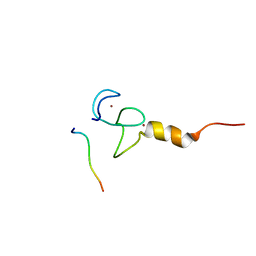 | | Structural basis of RACK7 PHD to read a pediatric glioblastoma-associated histone mutation H3.3G34R | | Descriptor: | Peptide from Histone H3.3, Protein kinase C-binding protein 1, ZINC ION | | Authors: | Lan, W.X, Li, Z, Jiao, F.F, Wang, C.X, Guo, R, Cao, C.Y. | | Deposit date: | 2020-08-28 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of RACK7 PHD domain to read a pediatric glioblastoma‐associated histone mutation H3.3G34R
Chin.J.Chem., 2021
|
|
7D0X
 
 | |
7D0Y
 
 | |
7D0Z
 
 | |
7D11
 
 | |
7D12
 
 | | NMR solution structures of CAG RNA-DB213 binding complex | | Descriptor: | N'-{(Z)-amino[4-(amino{[3-(dimethylammonio)propyl]iminio}methyl)phenyl]methylidene}-N,N-dimethylpropane-1,3-diaminium, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*CP*UP*UP*CP*GP*GP*CP*AP*GP*CP*AP*GP*C)-3'), SODIUM ION | | Authors: | Chan, H.Y.E, Guo, P. | | Deposit date: | 2020-09-12 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | CAG RNAs induce DNA damage and apoptosis by silencing NUDT16 expression in polyglutamine degeneration.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7D13
 
 | | NERP-2 in a HFIP solution | | Descriptor: | Neurosecretory protein VGF | | Authors: | Park, O, Cheong, C, Jeon, Y. | | Deposit date: | 2020-09-13 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of neuroendocrine regulatory peptide-2 in membrane-mimicking environments.
Peptide Science, 2020
|
|
7D16
 
 | | NERP-2 in a DPC solution | | Descriptor: | Neurosecretory protein VGF | | Authors: | Park, O, Cheong, C, Jeon, Y. | | Deposit date: | 2020-09-13 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of neuroendocrine regulatory peptide-2 in membrane-mimicking environments.
Peptide Science, 2020
|
|
