6TJ3
 
 | |
6TKT
 
 | |
6TL0
 
 | | Solution structure and 1H, 13C and 15N chemical shift assignments for the complex of VPS29 with VARP 687-747 | | Descriptor: | Ankyrin repeat domain-containing protein 27, Vacuolar protein sorting-associated protein 29, ZINC ION | | Authors: | Owen, D.J, Neuhaus, D, Yang, J.-C, Crawley-Snowdon, H. | | Deposit date: | 2019-11-29 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism and evolution of the Zn-fingernail required for interaction of VARP with VPS29.
Nat Commun, 11, 2020
|
|
6TO6
 
 | | Solution structure of the modulator of repression (MOR) of the temperate bacteriophage TP901-1 from Lactococcus lactis | | Descriptor: | MOR | | Authors: | Rasmussen, K.K, Blackledge, M, Herrmann, T, Lo Leggio, L, Jensen, M.R. | | Deposit date: | 2019-12-11 | | Release date: | 2020-08-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Revealing the mechanism of repressor inactivation during switching of a temperate bacteriophage.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TOB
 
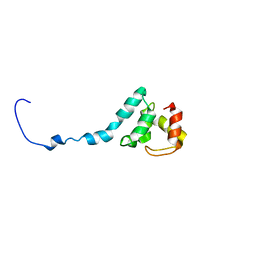 | |
6TPB
 
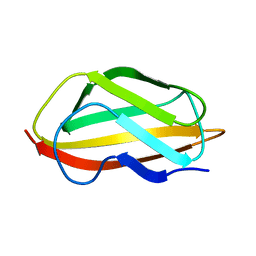 | | NMR structure of the apo-form of Pseudomonas fluorescens CopC | | Descriptor: | Putative copper resistance protein | | Authors: | Persson, K.C, Mayzel, M, Karlsson, B.G, Peciulyte, A, Olsson, L, Wittung Stafshede, P, Salomon Johansen, K, Horvath, I. | | Deposit date: | 2019-12-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Pseudomonas fluorescens CopC
To Be Published
|
|
6TPH
 
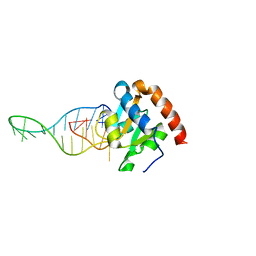 | |
6TR0
 
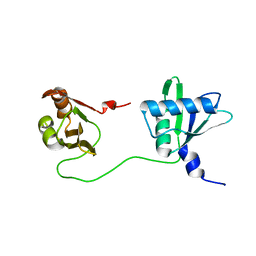 | | Solution structure of U2AF2 RRM1,2 | | Descriptor: | Splicing factor U2AF 65 kDa subunit | | Authors: | Kang, H.-S, Sattler, M. | | Deposit date: | 2019-12-17 | | Release date: | 2020-05-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | An autoinhibitory intramolecular interaction proof-reads RNA recognition by the essential splicing factor U2AF2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TR2
 
 | |
6TR8
 
 | | Corynebacterium diphtheriae methionine sulfoxide reductase B (MsrB) solution structure - reduced form | | Descriptor: | Peptide-methionine (R)-S-oxide reductase, ZINC ION | | Authors: | Volkov, A.N, Tossounian, M.A, Buts, L, Messens, J. | | Deposit date: | 2019-12-18 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Methionine sulfoxide reductase B fromCorynebacterium diphtheriaecatalyzes sulfoxide reduction via an intramolecular disulfide cascade.
J.Biol.Chem., 295, 2020
|
|
6TRM
 
 | | Solution structure of the antifungal protein PAFC | | Descriptor: | Pc21g12970 protein | | Authors: | Czajlik, A, Holzknecht, J, Marx, F, Batta, G. | | Deposit date: | 2019-12-19 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Dynamics, and New Antifungal Aspects of the Cysteine-Rich Miniprotein PAFC.
Int J Mol Sci, 22, 2021
|
|
6TRP
 
 | | Solution Structure of Docking Domain Complex of Pax NRPS: PaxC NDD - PaxB CDD | | Descriptor: | Peptide synthetase XpsB,Peptide synthetase XpsB | | Authors: | Watzel, J, Hacker, C, Duchardt-Ferner, E, Bode, H.B, Woehnert, J. | | Deposit date: | 2019-12-19 | | Release date: | 2020-08-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A New Docking Domain Type in the Peptide-Antimicrobial-Xenorhabdus Peptide Producing Nonribosomal Peptide Synthetase fromXenorhabdus bovienii.
Acs Chem.Biol., 15, 2020
|
|
6TT6
 
 | |
6TT8
 
 | | Haddock model of NDM-1/morin complex | | Descriptor: | 2-[2,4-bis(oxidanyl)phenyl]-3,5,7-tris(oxidanyl)chromen-4-one, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | Deposit date: | 2019-12-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
6TTA
 
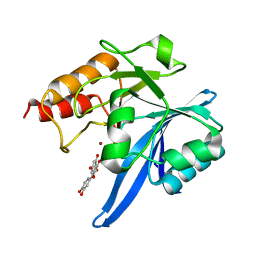 | | Haddock model of NDM-1/quercetin complex | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | Deposit date: | 2019-12-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
6TTC
 
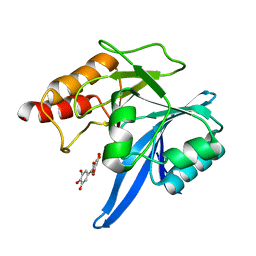 | | Haddock model of NDM-1/myricetin complex | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | Deposit date: | 2019-12-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
6TUB
 
 | | Beta-endorphin amyloid fibril | | Descriptor: | Beta-endorphin | | Authors: | Verasdonck, J, Seuring, C, Gath, J, Ghosh, D, Nespovitaya, N, Waelti, M.A, Maji, S, Cadalbert, R, Boeckmann, A, Guentert, P, Meier, B.H, Riek, R. | | Deposit date: | 2020-01-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The three-dimensional structure of human beta-endorphin amyloid fibrils.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6TV5
 
 | |
6TVJ
 
 | |
6TVM
 
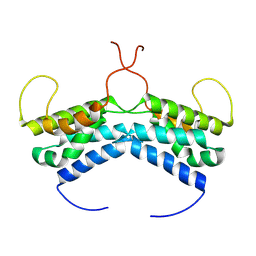 | | LEDGF/p75 dimer (residues 345-467) | | Descriptor: | PC4 and SFRS1-interacting protein | | Authors: | Lux, V, Veverka, V. | | Deposit date: | 2020-01-10 | | Release date: | 2020-09-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Molecular Mechanism of LEDGF/p75 Dimerization.
Structure, 28, 2020
|
|
6TWE
 
 | |
6TWG
 
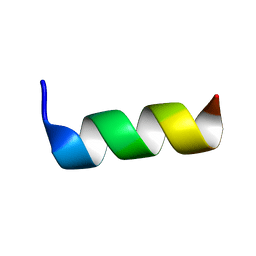 | | Solution structure of antimicrobial peptide, crabrolin Plus in the presence of Lipopolysaccharide | | Descriptor: | Crabrolin Plus, mutant of Crabrolin peptide | | Authors: | Cantini, F, Bouchemal, N, Savarin, P, Sette, M. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Effect of positive charges in the structural interaction of crabrolin isoforms with lipopolysaccharide.
J.Pept.Sci., 26, 2020
|
|
6TWR
 
 | |
6TXT
 
 | |
6TZE
 
 | | Human CstF-64 RRM mutant - D50A | | Descriptor: | Cleavage stimulation factor subunit 2 | | Authors: | Latham, M.P, Masoumzadeh, E. | | Deposit date: | 2019-08-12 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A missense mutation in the CSTF2 gene that impairs the function of the RNA recognition motif and causes defects in 3' end processing is associated with intellectual disability in humans.
Nucleic Acids Res., 48, 2020
|
|
