6QTF
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring B, major conformer | | Descriptor: | DCY-LEU-GLY-ALA-THR | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-02-25 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6QVW
 
 | | Solution structure of the free FOXO1 DNA binding domain | | Descriptor: | Forkhead box protein O1 | | Authors: | Psenakova, K, Obsil, T, Veverka, V, Obsilova, V, Kohoutova, K. | | Deposit date: | 2019-03-05 | | Release date: | 2019-09-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Forkhead Domains of FOXO Transcription Factors Differ in both Overall Conformation and Dynamics.
Cells, 8, 2019
|
|
6QWR
 
 | |
6QXB
 
 | | NMR structure of peptide 7, characterized by a cis-4-amino-Pro residue, with a significant lower MIC on E. coli | | Descriptor: | PHE-VAL-CAP-TRP-PHE-SER-LYS-PHE-LEU-GLY-ARG-ILE-LEU-NH2 | | Authors: | Brancaccio, D, Carotenuto, A, Merlino, F, Grieco, P, Novellino, E. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-29 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The Outcomes of Decorated Prolines in the Discovery of Antimicrobial Peptides from Temporin-L.
Chemmedchem, 14, 2019
|
|
6QXC
 
 | | NMR structure of peptide 8, characterized by a trans-4-cyclohexyl-Pro, with a dramatic reduction in activity on E. coli ATCC and lost effect on P. aeruginosa. | | Descriptor: | PHE-VAL-TCP-TRP-PHE-SER-LYS-PHE-LEU-GLY-ARG-ILE-LEU-NH2 | | Authors: | Brancaccio, D, Carotenuto, A, Merlino, F, Grieco, P, Novellino, E. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-29 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The Outcomes of Decorated Prolines in the Discovery of Antimicrobial Peptides from Temporin-L.
Chemmedchem, 14, 2019
|
|
6QXZ
 
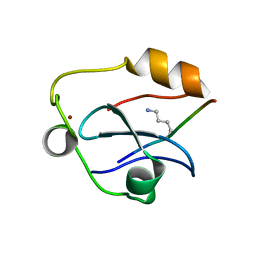 | | Solution structure of the ASHH2 CW domain with the N-terminal histone H3 tail mimicking peptide monomethylated on lysine 4 | | Descriptor: | ALA-ARG-THR-MLZ-GLN-THR-ALA-ARG-TYR, Histone-lysine N-methyltransferase ASHH2, ZINC ION | | Authors: | Dobrovolska, O, Madeleine, N, Teigen, K, Halskau, O, Bril'kov, M. | | Deposit date: | 2019-03-08 | | Release date: | 2019-12-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Arabidopsis (ASHH2) CW domain binds monomethylated K4 of the histone H3 tail through conformational selection.
Febs J., 287, 2020
|
|
6QYR
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring B, minor conformer | | Descriptor: | DAL-LEU-GLY-CYS-THR | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6QYS
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Nisin Ring B | | Descriptor: | DBB-PRO-GLY-CYS-LYS | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6QYT
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring A truncated analogue | | Descriptor: | DAL-LEU-SER-LEU-CYS-ALA | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6QYU
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring A | | Descriptor: | PHE-DHA-DAL-LEU-DHA-LEU-CYS-ALA | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6QYV
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring A (Ser2, Ala5, Ala8) analogue | | Descriptor: | PHE-SER-DAL-LEU-ALA-LEU-CYS-ALA | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6QYW
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - nisin ring A | | Descriptor: | ILE-DBU-DAL-ILE-DHA-LEU-CYS-ALA | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2019-10-02 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
6R0J
 
 | |
6R14
 
 | | Structure of kiteplatinated dsDNA | | Descriptor: | Kiteplatin, Kiteplatinated DNA oligomer, chain A, ... | | Authors: | Margiotta, N, Papadia, P, Kubicek, K, Krejcikova, M, Gkionis, K, Sponer, J. | | Deposit date: | 2019-03-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of kiteplatinated DNA
To Be Published
|
|
6R1V
 
 | |
6R28
 
 | |
6R2X
 
 | | NMR structure of Chromogranin A (F39-D63) | | Descriptor: | Chromogranin-A | | Authors: | Nardelli, F, Quilici, G, Ghitti, M, Curnis, F, Gori, A, Berardi, A, Corti, A, Musco, G. | | Deposit date: | 2019-03-19 | | Release date: | 2019-12-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A stapled chromogranin A-derived peptide is a potent dual ligand for integrins alpha v beta 6 and alpha v beta 8.
Chem.Commun.(Camb.), 55, 2019
|
|
6R3C
 
 | |
6R5G
 
 | |
6R8E
 
 | | SC14 G-hairpin | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*GP*TP*GP*GP*GP*TP*GP*TP*GP*T)-3') | | Authors: | Lenarcic Zivkovic, M, Trantirek, L, Plavec, J. | | Deposit date: | 2019-04-01 | | Release date: | 2021-02-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Insight into formation propensity of pseudocircular DNA G-hairpins.
Nucleic Acids Res., 49, 2021
|
|
6R8N
 
 | | STRUCTURE DETERMINATION OF THE TETRAHEDRAL AMINOPEPTIDASE TET2 FROM P. HORIKOSHII BY USE OF COMBINED SOLID-STATE NMR, SOLUTION-STATE NMR AND EM DATA 4.1 A, FOLLOWED BY REAL_SPACE_REFINEMENT AT 4.1 A | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Colletier, J.-P, Gauto, D, Estrozi, L, Favier, A, Effantin, G, Schoehn, G, Boisbouvier, J, Schanda, P. | | Deposit date: | 2019-04-02 | | Release date: | 2019-08-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
6R95
 
 | |
6R96
 
 | |
6R9K
 
 | |
6R9L
 
 | |
