5O9B
 
 | |
5OAO
 
 | |
5OAP
 
 | |
5OAY
 
 | |
5OB4
 
 | |
5OBN
 
 | | NMR solution structure of U11/U12 65K protein's C-terminal RRM domain (381-516) | | Descriptor: | RNA-binding protein 40 | | Authors: | Norppa, A.J, Kauppala, T.M, Heikkinen, H.A, Verma, B, Iwai, H, Frilander, M.J. | | Deposit date: | 2017-06-28 | | Release date: | 2018-01-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Mutations in the U11/U12-65K protein associated with isolated growth hormone deficiency lead to structural destabilization and impaired binding of U12 snRNA.
RNA, 24, 2018
|
|
5OCZ
 
 | | Free DNA_hairpin polyamides studies | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*GP*TP*AP*CP*AP*TP*CP*G)-3') | | Authors: | Padroni, G, Parkinson, J, Burley, G.A. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA duplex distortion induced by thiazole-containing hairpin polyamides.
Nucleic Acids Res., 46, 2018
|
|
5ODD
 
 | | HUMAN MED26 N-TERMINAL DOMAIN (1-92) | | Descriptor: | Mediator of RNA polymerase II transcription subunit 26 | | Authors: | Lens, Z, Cantrelle, F.-X, Perruzini, R, Dewitte, F, Hanoulle, X, Villeret, V, Verger, A, Landrieu, I. | | Deposit date: | 2017-07-05 | | Release date: | 2017-11-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | 1H, 15N and 13C assignments of the N-terminal domain of the Mediator complex subunit MED26.
Biomol.Nmr Assign., 10, 2016
|
|
5ODF
 
 | |
5ODM
 
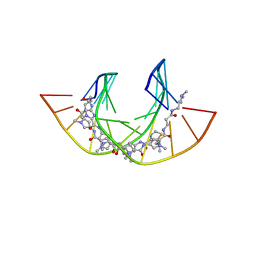 | | NtiPr polyamide in complex with 5'CGATGTACTACG3 | | Descriptor: | 3-(3-azaniumylpropanoylamino)propyl-dimethyl-azanium, 4-AMINO-(1-METHYLIMIDAZOLE)-2-CARBOXYLIC ACID, 4-AMINO-(1-METHYLPYRROLE)-2-CARBOXYLIC ACID, ... | | Authors: | Padroni, G, Parkinson, J, Burley, G.A. | | Deposit date: | 2017-07-06 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA duplex distortion induced by thiazole-containing hairpin polyamides.
Nucleic Acids Res., 46, 2018
|
|
5OE1
 
 | | Im polyamide in complex with 5'CGATGTACATCG3'- hairpin polyamides studies | | Descriptor: | 1-methylimidazole-2-carboxylic acid, 3-(3-azaniumylpropanoylamino)propyl-dimethyl-azanium, 4-AMINO-(1-METHYLIMIDAZOLE)-2-CARBOXYLIC ACID, ... | | Authors: | Padroni, G, Parkinson, J, Burley, G.A. | | Deposit date: | 2017-07-07 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA duplex distortion induced by thiazole-containing hairpin polyamides.
Nucleic Acids Res., 46, 2018
|
|
5OEK
 
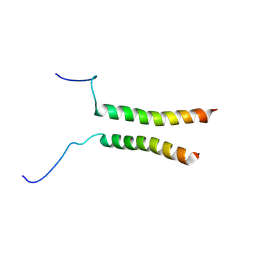 | | Putative active dimeric state of GHR transmembrane domain | | Descriptor: | Growth hormone receptor | | Authors: | Lesovoy, D.M, Bocharov, E.V, Bocharova, O.V, Urban, A.S, Arseniev, A.S. | | Deposit date: | 2017-07-08 | | Release date: | 2018-04-11 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the signal transduction via transmembrane domain of the human growth hormone receptor.
Biochim. Biophys. Acta, 1862, 2018
|
|
5OEO
 
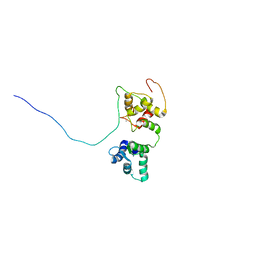 | | Solution structure of the complex of TRPV5(655-725) with a Calmodulin E32Q/E68Q double mutant | | Descriptor: | CALCIUM ION, Calmodulin-1, Transient receptor potential cation channel subfamily V member 5 | | Authors: | Vuister, G.W, Bokhovchuk, F.M, Bate, N, Kovalevskaya, N, Goult, B.T, Spronk, C.A.E.M. | | Deposit date: | 2017-07-09 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structural Basis of Calcium-Dependent Inactivation of the Transient Receptor Potential Vanilloid 5 Channel.
Biochemistry, 57, 2018
|
|
5OGA
 
 | | Structure of minimal i-motif domain | | Descriptor: | DNA (5'-D(*TP*(DCP)P*GP*TP*TP*CP*(DCP)P*GP*TP*TP*TP*TP*TP*CP*GP*TP*TP*CP*CP*GP*T)-3') | | Authors: | Mir, B, Serrano, I, Buitrago, D, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2017-07-12 | | Release date: | 2017-11-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Prevalent Sequences in the Human Genome Can Form Mini i-Motif Structures at Physiological pH.
J. Am. Chem. Soc., 139, 2017
|
|
5OGU
 
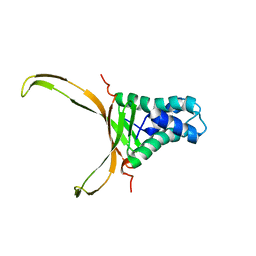 | | Structure of DNA-binding HU protein from micoplasma Spiroplasma melliferum | | Descriptor: | DNA-binding protein | | Authors: | Altukhov, D.A, Talyzina, A.A, Agapova, Y.K, Vlaskina, A.V, Korzhenevskiy, D.A, Bocharov, E.V, Rakitina, T.V, Timofeev, V.I. | | Deposit date: | 2017-07-13 | | Release date: | 2017-08-23 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural plasticity and thermal stability of the histone-like protein from Spiroplasma melliferum are due to phenylalanine insertions into the conservative scaffold.
J.Biomol.Struct.Dyn., 36, 2018
|
|
5OHD
 
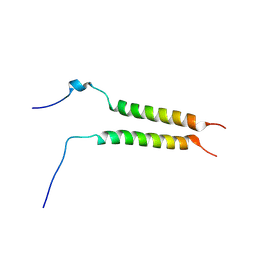 | | Putative inactive (dormant) dimeric state of GHR transmembrane domain | | Descriptor: | Growth hormone receptor | | Authors: | Lesovoy, D.M, Bocharov, E.V, Bocharova, O.V, Urban, A.S, Arseniev, A.S. | | Deposit date: | 2017-07-15 | | Release date: | 2018-04-11 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the signal transduction via transmembrane domain of the human growth hormone receptor.
Biochim. Biophys. Acta, 1862, 2018
|
|
5OJT
 
 | |
5OLF
 
 | |
5OMZ
 
 | |
5OPH
 
 | |
5OQK
 
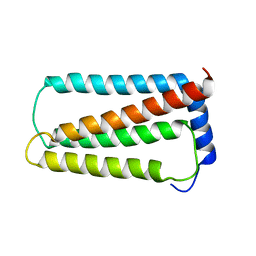 | | Solution NMR structure of truncated, human Hv1/VSOP (Voltage-gated proton channel) | | Descriptor: | Voltage-gated hydrogen channel 1 | | Authors: | Bayrhuber, M, Maslennikov, I, Kwiatowski, W, Sobol, A, Wierschem, C, Eichmann, C, Riek, R. | | Deposit date: | 2017-08-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure and Functional Behavior of the Human Proton Channel.
Biochemistry, 58, 2019
|
|
5OQS
 
 | | Solution structure of antifungal protein NFAP | | Descriptor: | NFAP | | Authors: | Hajdu, D, Czajlik, A, Marx, F, Galgoczy, L, Batta, G. | | Deposit date: | 2017-08-14 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and novel insights into phylogeny and mode of action of the Neosartorya (Aspergillus) fischeri antifungal protein (NFAP).
Int.J.Biol.Macromol., 129, 2019
|
|
5OR0
 
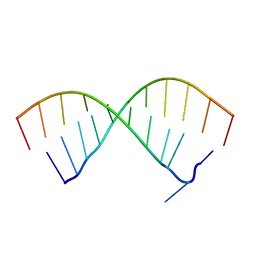 | |
5OR5
 
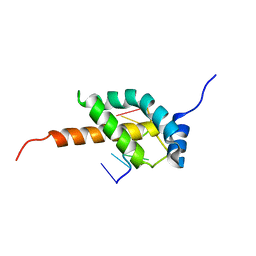 | |
5OUN
 
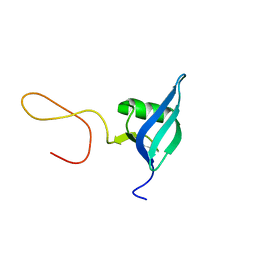 | | NMR solution structure of the external DII domain of Rvb2 from Saccharomyces cerevisiae | | Descriptor: | RuvB-like protein 2 | | Authors: | Rouillon, C, Bragantini, B, Charpentier, B, Manival, X, Quinternet, M. | | Deposit date: | 2017-08-24 | | Release date: | 2018-03-28 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | NMR assignment and solution structure of the external DII domain of the yeast Rvb2 protein.
Biomol NMR Assign, 12, 2018
|
|
