2RT5
 
 | |
2RT6
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for PriC N-terminal domain | | Descriptor: | Primosomal replication protein N'' | | Authors: | Aramaki, T, Abe, Y, Katayama, T, Ueda, T. | | Deposit date: | 2013-04-24 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of a replication restart primosome factor, PriC, in Escherichia coli.
Protein Sci., 22, 2013
|
|
2RT8
 
 | | Structure of metallo-dna in solution | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*TP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*TP*TP*CP*GP*CP*G)-3'), MERCURY (II) ION | | Authors: | Yamaguchi, H, Sebera, J, Kondo, J, Oda, S, Komuro, T, Kawamura, T, Dairaku, T, Kondo, Y, Okamoto, I, Ono, A, Burda, J.V, Kojima, C, Sychrovsky, V, Tanaka, Y. | | Deposit date: | 2013-06-18 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of metallo-DNA with consecutive thymine-HgII-thymine base pairs explains positive entropy for the metallo base pair formation.
Nucleic Acids Res., 42, 2014
|
|
2RT9
 
 | | Solution structure of a regulatory domain of meiosis inhibitor | | Descriptor: | F-box only protein 43, ZINC ION | | Authors: | Shoji, S, Muto, Y, Ikeda, M, He, F, Tsuda, K, Ohsawa, N, Akasaka, R, Terada, T, Wakiyama, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The zinc-binding region (ZBR) fragment of Emi2 can inhibit APC/C by targeting its association with the coactivator Cdc20 and UBE2C-mediated ubiquitylation
FEBS Open Bio, 4, 2014
|
|
2RTS
 
 | | Chitin binding domain1 | | Descriptor: | chitinase | | Authors: | Uegaki, K. | | Deposit date: | 2013-08-19 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the chitin-binding domain 1 (ChBD1) of a hyperthermophilic chitinase from Pyrococcus furiosus.
J.Biochem., 155, 2014
|
|
2RTT
 
 | | Solution structure of the chitin-binding domain of Chi18aC from Streptomyces coelicolor | | Descriptor: | ChiC | | Authors: | Okumura, A, Uemura, M, Yamada, N, Chikaishi, E, Takai, T, Yoshio, S, Akagi, K, Morita, J, Lee, Y, Yokogawa, D, Suzuki, K, Watanabe, T, Ikegami, T. | | Deposit date: | 2013-08-26 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Chitin-binding domain of chitinase Chi18aC from Streptomyces coelicolor
To be Published
|
|
2RTU
 
 | | Solution structure of oxidized human HMGB1 A box | | Descriptor: | High mobility group protein B1 | | Authors: | Jing, W, Tochio, N, Tate, S. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Redox-sensitive structural change in the A-domain of HMGB1 and its implication for the binding to cisplatin modified DNA.
Biochem.Biophys.Res.Commun., 441, 2013
|
|
2RTV
 
 | | Tachyplesin I in water | | Descriptor: | Tachyplesin-1 | | Authors: | Kushibiki, T, Kamiya, M, Aizawa, T, Kumaki, Y, Kikukawa, T, Mizuguchi, M, Demura, M, Kawabata, S.I, Kawano, K. | | Deposit date: | 2013-09-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Interaction between tachyplesin I, an antimicrobial peptide derived from horseshoe crab, and lipopolysaccharide.
Biochim.Biophys.Acta, 1844, 2014
|
|
2RTX
 
 | | Solution structure of the GGQ domain of YaeJ protein from Escherichia coli | | Descriptor: | Peptidyl-tRNA hydrolase YaeJ | | Authors: | Nameki, N, Enomoto, M, Kogure, H, Tochio, N, Guntert, P. | | Deposit date: | 2013-09-21 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification of residues required for stalled-ribosome rescue in the codon-independent release factor YaeJ
Nucleic Acids Res., 42, 2014
|
|
2RTY
 
 | | Solution structure of navitoxin | | Descriptor: | navitoxin | | Authors: | Umetsu, Y, Ohki, S. | | Deposit date: | 2013-10-16 | | Release date: | 2014-04-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Experimental conversion of a defensin into a neurotoxin: implications for origin of toxic function
MOL.BIOL.EVOL., 31, 2014
|
|
2RTZ
 
 | | Solution structure of MarkTX-7 | | Descriptor: | Potassium channel blocker MarkTX-7 | | Authors: | Umetsu, Y, Ohki, S. | | Deposit date: | 2013-10-17 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of MarkTX-7
To be Published
|
|
2RU0
 
 | |
2RU1
 
 | | Solution structure of esf3 | | Descriptor: | Uncharacterized protein | | Authors: | Umetsu, Y, Mori, M, Ohki, S. | | Deposit date: | 2013-11-06 | | Release date: | 2014-04-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Central cell-derived peptides regulate early embryo patterning in flowering plants
Science, 344, 2014
|
|
2RU2
 
 | | NMR solution structure of [G5,T7,S9]-oxytocin | | Descriptor: | [G5,T7,S9]-oxytocin | | Authors: | Harvey, P, Craik, D. | | Deposit date: | 2013-11-08 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Oxytocic plant cyclotides as templates for peptide G protein-coupled receptor ligand design.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2RU3
 
 | | Solution structure of c.elegans SUP-12 RRM in complex with RNA | | Descriptor: | Protein SUP-12, isoform a, RNA (5'-R(*GP*UP*GP*UP*GP*C)-3') | | Authors: | Takahashi, M, Kuwasako, K, Unzai, S, Tsuda, K, Yoshikawa, S, He, F, Kobayashi, N, Guntert, P, Shirouzu, M, Ito, T, Tanaka, A, Yokoyama, S, Hagiwara, M, Kuroyanagi, H, Muto, Y. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RBFOX and SUP-12 sandwich a G base to cooperatively regulate tissue-specific splicing
Nat.Struct.Mol.Biol., 21, 2014
|
|
2RU4
 
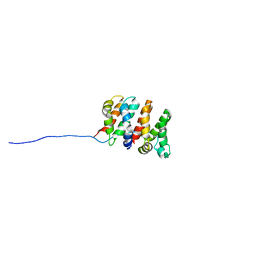 | | Designed Armadillo Repeat Protein Self-ASsembled Complex (YIIM2-MAII) | | Descriptor: | Armadillo Repeat Protein, C-terminal fragment, MAII, ... | | Authors: | Zerbe, O, Christen, M.T, Plueckthun, A, Watson, R.P. | | Deposit date: | 2013-11-22 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Spontaneous self-assembly of engineered armadillo repeat protein fragments into a folded structure
Structure, 22, 2014
|
|
2RU5
 
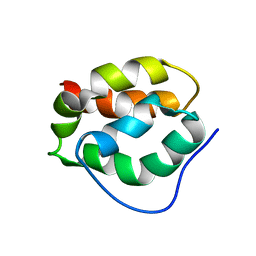 | |
2RU6
 
 | | The pure alternative state of ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Kitazawa, S, Kameda, T, Kumo, A, Utsumi, M, Baxter, N, Kato, K, Williamson, M.P, Kitahara, R. | | Deposit date: | 2013-12-04 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Close Identity between Alternatively Folded State N2 of Ubiquitin and the Conformation of the Protein Bound to the Ubiquitin-Activating Enzyme
Biochemistry, 53, 2014
|
|
2RU7
 
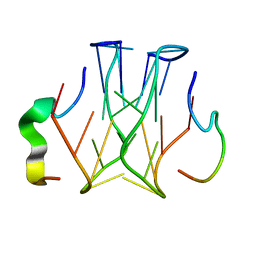 | | Refined structure of RNA aptamer in complex with the partial binding peptide of prion protein | | Descriptor: | P16 peptide from Major prion protein, RNA_(5'-R(*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*A)-3') | | Authors: | Hayashi, T, Oshima, H, Mashima, T, Nagata, T, Katahira, M, Kinoshita, M. | | Deposit date: | 2013-12-24 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Binding of an RNA aptamer and a partial peptide of a prion protein: crucial importance of water entropy in molecular recognition.
Nucleic Acids Res., 42, 2014
|
|
2RU8
 
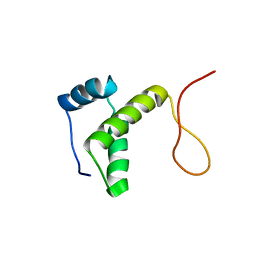 | | DnaT C-terminal domain | | Descriptor: | Primosomal protein 1 | | Authors: | Abe, Y, Tani, J, Fujiyama, S, Urabe, M, Sato, K, Aramaki, T, Katayama, T, Ueda, T. | | Deposit date: | 2014-01-29 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and mechanism of the primosome protein DnaT-functional structures for homotrimerization, dissociation of ssDNA from the PriB·ssDNA complex, and formation of the DnaT·ssDNA complex.
Febs J., 281, 2014
|
|
2RU9
 
 | |
2RUC
 
 | |
2RUD
 
 | |
2RUE
 
 | |
2RUF
 
 | |
