6V10
 
 | | genome-containing AAV8 particles | | Descriptor: | Capsid protein, DNA (5'-D(*CP*A)-3') | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2019-11-19 | | Release date: | 2019-12-11 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Comparative Analysis of the Capsid Structures of AAVrh.10, AAVrh.39, and AAV8.
J.Virol., 94, 2020
|
|
6V12
 
 | | Empty AAV8 particles | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2019-11-19 | | Release date: | 2019-12-11 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Comparative Analysis of the Capsid Structures of AAVrh.10, AAVrh.39, and AAV8.
J.Virol., 94, 2020
|
|
6V1G
 
 | | Genome-containing AAVrh.10 | | Descriptor: | Capsid protein VP1, DNA (5'-D(*CP*A)-3') | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2019-11-20 | | Release date: | 2019-12-11 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Comparative Analysis of the Capsid Structures of AAVrh.10, AAVrh.39, and AAV8.
J.Virol., 94, 2020
|
|
6V1T
 
 | | Empty AAVrh.39 particle | | Descriptor: | Capsid protein VP1 | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2019-11-21 | | Release date: | 2019-12-11 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Comparative Analysis of the Capsid Structures of AAVrh.10, AAVrh.39, and AAV8.
J.Virol., 94, 2020
|
|
6V1Z
 
 | | genome-containing AAVrh.39 particles | | Descriptor: | Capsid protein VP1, DNA (5'-D(*CP*A)-3') | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2019-11-21 | | Release date: | 2019-12-11 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Comparative Analysis of the Capsid Structures of AAVrh.10, AAVrh.39, and AAV8.
J.Virol., 94, 2020
|
|
6V39
 
 | |
6V3A
 
 | |
6V3B
 
 | |
6V3D
 
 | |
6V3E
 
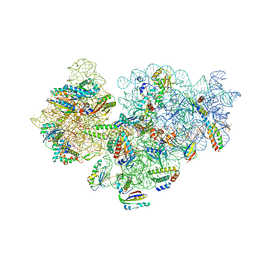 | |
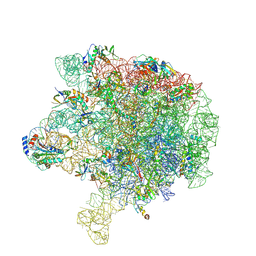
6V41
 
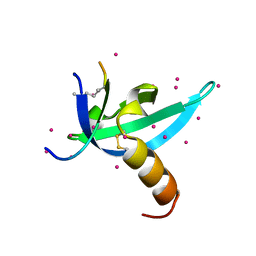 | | crystal structure of CDY1 chromodomain bound to H3K9me3 | | Descriptor: | Histone H3.1 Peptide, Testis-specific chromodomain protein Y 1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-27 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V6S
 
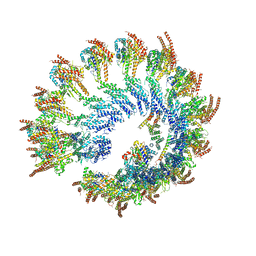 | | Structure of the native human gamma-tubulin ring complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Gamma-tubulin complex component 2, ... | | Authors: | Wieczorek, M, Urnavicius, L, Ti, S, Molloy, K.R, Chait, B.T, Kapoor, T.M. | | Deposit date: | 2019-12-05 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Asymmetric Molecular Architecture of the Human gamma-Tubulin Ring Complex.
Cell, 180, 2020
|
|
6V8I
 
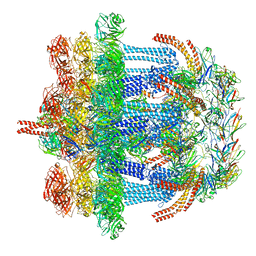 | |
6V8W
 
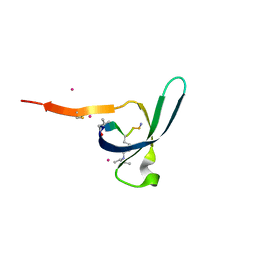 | | CDYL2 chromodomain in complex with a synthetic peptide | | Descriptor: | Chromodomain Y-like protein 2, IVA-PHE-ALA-PHE-5T3-SER-NH2, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, James, L.I, Lamb, K.N, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V9T
 
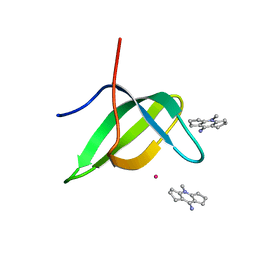 | | Tudor domain of TDRD3 in complex with a small molecule | | Descriptor: | 4-methyl-2,3,4,5,6,7-hexahydrodicyclopenta[b,e]pyridin-8(1H)-imine, Tudor domain-containing protein 3, UNKNOWN ATOM OR ION | | Authors: | Li, W, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-16 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
6VE7
 
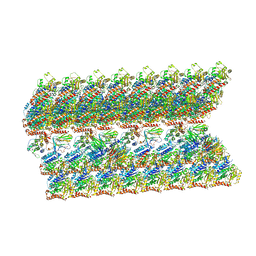 | | The inner junction complex of Chlamydomonas reinhardtii doublet microtubule | | Descriptor: | Cilia- and flagella-associated protein 20, FAP276, FAP52, ... | | Authors: | Khalifa, A.A.Z, Ichikawa, M, Bui, K.H. | | Deposit date: | 2019-12-30 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The inner junction complex of the cilia is an interaction hub that involves tubulin post-translational modifications.
Elife, 9, 2020
|
|
6VGS
 
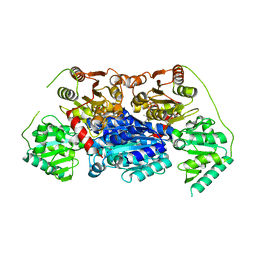 | | Alpha-ketoisovalerate decarboxylase (KivD) from Lactococcus lactis, thermostable mutant | | Descriptor: | Alpha-keto acid decarboxylase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Chan, S, Korman, T.P, Sawaya, M.R, Bowie, J.U. | | Deposit date: | 2020-01-08 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isobutanol production freed from biological limits using synthetic biochemistry.
Nat Commun, 11, 2020
|
|
6VHO
 
 | |
6VLZ
 
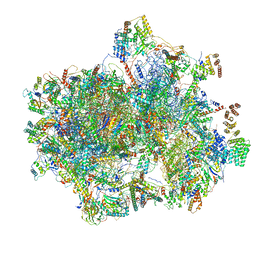 | | Structure of the human mitochondrial ribosome-EF-G1 complex (ClassI) | | Descriptor: | 12s rRNA, 16s rRNA, 28S ribosomal protein S10, ... | | Authors: | Sharma, M.R, Koripella, R.K, Agrawal, R.K. | | Deposit date: | 2020-01-27 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of the human mitochondrial ribosome bound to EF-G1 reveal distinct features of mitochondrial translation elongation.
Nat Commun, 11, 2020
|
|
6VMI
 
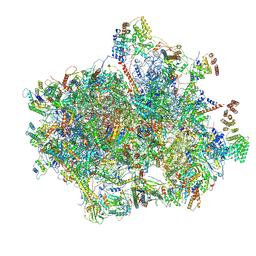 | | Structure of the human mitochondrial ribosome-EF-G1 complex (ClassIII) | | Descriptor: | 12s rRNA, 16s rRNA, 28S ribosomal protein S10, ... | | Authors: | Sharma, M.R, Koripella, R.K, Agrawal, R.K. | | Deposit date: | 2020-01-28 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structures of the human mitochondrial ribosome bound to EF-G1 reveal distinct features of mitochondrial translation elongation.
Nat Commun, 11, 2020
|
|
6VU3
 
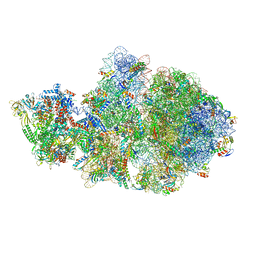 | | Cryo-EM structure of Escherichia coli transcription-translation complex A (TTC-A) containing mRNA with a 12 nt long spacer | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R. | | Deposit date: | 2020-02-14 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6VVH
 
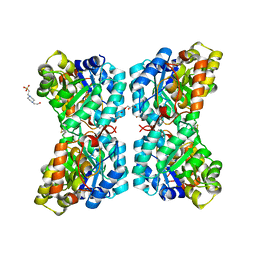 | |
6VVI
 
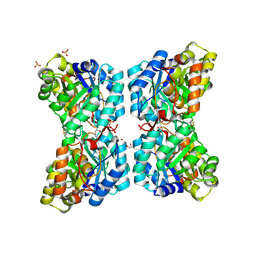 | |
6VWL
 
 | | 70S ribosome bound to HIV frameshifting stem-loop (FSS) and P/E tRNA (rotated conformation) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loerch, S, Bao, C, Ling, C, Korostelev, A.A, Grigorieff, N, Ermolenko, D.M. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | mRNA stem-loops can pause the ribosome by hindering A-site tRNA binding.
Elife, 9, 2020
|
|
6VWM
 
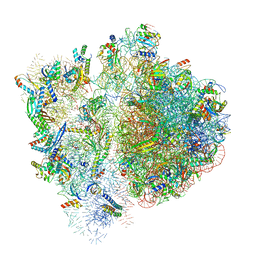 | | 70S ribosome bound to HIV frameshifting stem-loop (FSS) and P-site tRNA (non-rotated conformation, Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loerch, S, Bao, C, Ling, C, Korostelev, A.A, Grigorieff, N, Ermolenko, D.M. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | mRNA stem-loops can pause the ribosome by hindering A-site tRNA binding.
Elife, 9, 2020
|
|







