6YTN
 
 | |
6YTP
 
 | | Structure of recombinant human beta-glucocerebrosidase in complex with azide tagged cyclophellitol epoxide inhibitor | | Descriptor: | (1~{S},2~{R},3~{R},4~{S},5~{S})-4-[[($l^{5}-azanylidyne-$l^{5}-azanyl)amino]methyl]cyclohexane-1,2,3,5-tetrol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2020-04-24 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, Synthesis and Structural Analysis of Glucocerebrosidase Imaging Agents.
Chemistry, 27, 2021
|
|
6YTR
 
 | | Structure of recombinant human beta-glucocerebrosidase in complex with cyclophellitol aziridine inhibitor | | Descriptor: | (1~{R},2~{S},3~{S},4~{S},5~{R},6~{R})-5-azanyl-6-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2020-04-24 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, Synthesis and Structural Analysis of Glucocerebrosidase Imaging Agents.
Chemistry, 27, 2021
|
|
6YUL
 
 | | CK2 alpha bound to Macrocycle | | Descriptor: | 7,10-Dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.12,6.017,20]docosa-1(20),2(22),3,5,14(21),15,18-heptaene-5-carboxylic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Kraemer, A, Hanke, T, Kurz, C, Celik, I, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-27 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of pyrazolo[1,5-a]pyrimidines lead to the identification of a highly selective casein kinase 2 inhibitor.
Eur.J.Med.Chem., 208, 2020
|
|
6YUM
 
 | | CK2 alpha bound to unclosed Macrocycle | | Descriptor: | 4-[5-[2-(2-hydroxyethyloxy)ethyl-[(2-methylpropan-2-yl)oxycarbonyl]amino]pyrazolo[1,5-a]pyrimidin-3-yl]-2-oxidanyl-benzoic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Kraemer, A, Hanke, T, Kurz, C, Celik, I, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-27 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Optimization of pyrazolo[1,5-a]pyrimidines lead to the identification of a highly selective casein kinase 2 inhibitor.
Eur.J.Med.Chem., 208, 2020
|
|
6YUT
 
 | | Structure of recombinant human beta-glucocerebrosidase in complex with N-acyl functionalised cyclophellitol aziridine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2020-04-27 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Design, Synthesis and Structural Analysis of Glucocerebrosidase Imaging Agents.
Chemistry, 27, 2021
|
|
6YV3
 
 | | Structure of recombinant human beta-glucocerebrosidase in complex with galacto-configured cyclophellitol aziridine inhibitor | | Descriptor: | (1S,2S,3S,4S,5R,6R)-5-amino-6-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2020-04-27 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis and Structural Analysis of Glucocerebrosidase Imaging Agents.
Chemistry, 27, 2021
|
|
6YVE
 
 | | Glycogen phosphorylase b in complex with pelargonidin 3-O-beta-D-glucoside | | Descriptor: | DIMETHYL SULFOXIDE, Glycogen phosphorylase, muscle form, ... | | Authors: | Drakou, C.E, Gardeli, C, Tsialtas, I, Alexopoulos, S, Mallouchos, A, Koulas, S, Tsagkarakou, A, Asimakopoulos, D, Leonidas, D.D, Psarra, A.M, Skamnaki, V.T. | | Deposit date: | 2020-04-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Affinity Crystallography Reveals Binding of Pomegranate Juice Anthocyanins at the Inhibitor Site of Glycogen Phosphorylase: The Contribution of a Sugar Moiety to Potency and Its Implications to the Binding Mode.
J.Agric.Food Chem., 68, 2020
|
|
6YVP
 
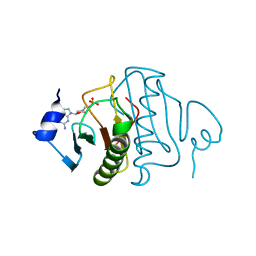 | | Human histidine triad nucleotide-binding protein 2 (hHINT2) complexed with dGMP and refined to 2.77 A | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, Histidine triad nucleotide-binding protein 2, mitochondrial | | Authors: | Dolot, R.D, Krakowiak, A, Nawrot, B.C. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Biochemical, crystallographic and biophysical characterization of histidine triad nucleotide-binding protein 2 with different ligands including a non-hydrolyzable analog of Ap4A.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
6YVR
 
 | | Crystal structure of the neurotensin receptor 1 in complex with the peptide full agonist NTS8-13 | | Descriptor: | Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin crystallisation chaperone, neurotensin NTS8-13 (full agonist), nonyl beta-D-glucopyranoside | | Authors: | Deluigi, M, Merklinger, L, Hilge, M, Ernst, P, Klipp, A, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-04-28 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.458 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6YW5
 
 | | The structure of the small subunit of the mitoribosome from Neurospora crassa | | Descriptor: | 16S rRNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Amunts, A, Itoh, Y, Naschberger, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi.
Nat Commun, 11, 2020
|
|
6YWE
 
 | | The structure of the mitoribosome from Neurospora crassa in the P/E tRNA bound state | | Descriptor: | 16S rRNA, 23S rRNA, 3-hydroxyisobutyryl-CoA hydrolase, ... | | Authors: | Amunts, A, Itoh, Y, Naschberger, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi.
Nat Commun, 11, 2020
|
|
6YWS
 
 | | The structure of the large subunit of the mitoribosome from Neurospora crassa | | Descriptor: | 50S ribosomal protein L14, 50S ribosomal protein L17, 50S ribosomal protein L24, ... | | Authors: | Amunts, A, Itoh, Y, Naschberger, A. | | Deposit date: | 2020-04-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi.
Nat Commun, 11, 2020
|
|
6YWV
 
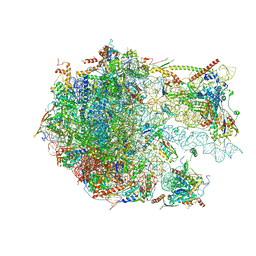 | | The structure of the Atp25 bound assembly intermediate of the mitoribosome from Neurospora crassa | | Descriptor: | 23 S rRNA, 50S ribosomal protein L14, 50S ribosomal protein L17, ... | | Authors: | Amunts, A, Itoh, Y, Naschberger, A. | | Deposit date: | 2020-04-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi.
Nat Commun, 11, 2020
|
|
6YWX
 
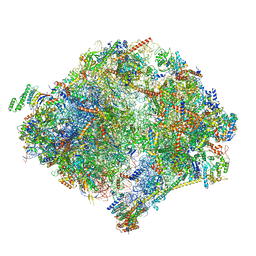 | | The structure of the mitoribosome from Neurospora crassa with tRNA bound to the E-site | | Descriptor: | 16S rRNA, 23S rRNA, 3-hydroxyisobutyryl-CoA hydrolase, ... | | Authors: | Amunts, A, Itoh, Y, Naschberger, A. | | Deposit date: | 2020-04-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi.
Nat Commun, 11, 2020
|
|
6YWY
 
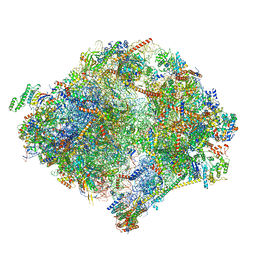 | | The structure of the mitoribosome from Neurospora crassa with bound tRNA at the P-site | | Descriptor: | 16S rRNA, 23S rRNA, 3-hydroxyisobutyryl-CoA hydrolase, ... | | Authors: | Amunts, A, Itoh, Y, Naschberger, A. | | Deposit date: | 2020-04-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi.
Nat Commun, 11, 2020
|
|
6YX7
 
 | | The high resolution structure of allophycocyanin from cyanobacterium Nostoc sp. WR13, the P21212 crystal form. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Patel, H.M, Roszak, A.W, Madamwar, D, Cogdell, R.J. | | Deposit date: | 2020-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | The high resolution structure of allophycocyanin from cyanobacterium Nostoc sp. WR13
To Be Published
|
|
6YX8
 
 | | The structure of allophycocyanin from cyanobacterium Nostoc sp. WR13, the C2221 crystal form. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Patel, H.M, Roszak, A.W, Madamwar, D, Cogdell, R.J. | | Deposit date: | 2020-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.831 Å) | | Cite: | The high resolution structure of allophycocyanin from cyanobacterium Nostoc sp. WR13
To Be Published
|
|
6YXL
 
 | | Crystal structure of ACPA F3 | | Descriptor: | ACPA F3 Fab fragment - heavy chain, ACPA F3 Fab fragment - light chain, GLYCEROL, ... | | Authors: | Ge, C, Holmdahl, R. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Surface Ig variable domain glycosylation affects autoantigen binding and acts as threshold for human autoreactive B cell activation.
Sci Adv, 8, 2022
|
|
6YXM
 
 | | Crystal structure of ACPA 1F2 in complex with CII-C-39-CIT | | Descriptor: | ACPA 1F2 Fab fragment - heavy chain, ACPA 1F2 Fab fragment - light chain, CII-C-39-CIT, ... | | Authors: | Ge, C, Holmdahl, R. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Surface Ig variable domain glycosylation affects autoantigen binding and acts as threshold for human autoreactive B cell activation.
Sci Adv, 8, 2022
|
|
6YXX
 
 | | State A of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | Descriptor: | 12S ribosomal RNA, 50S ribosomal protein L13, putative, ... | | Authors: | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2020-05-04 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
6YXY
 
 | | State B of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | Descriptor: | 12S ribosomal RNA, ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2020-05-04 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
6YXZ
 
 | |
6YY5
 
 | | Crystal structure of the ferric enterobactin receptor (PfeA) in complex with TCV_L5 | | Descriptor: | FE (III) ION, Ferric enterobactin receptor, ~{N}-[2-[[(2~{S})-3-[[(2~{S})-3-[[1-[2-[2-[2-[4-[4-[5-(acetamidomethyl)-2-oxidanylidene-1,3-oxazolidin-3-yl]-2-fluoranyl-phenyl]piperazin-1-yl]-2-oxidanylidene-ethoxy]ethoxy]ethyl]-1,2,3-triazol-4-yl]methylamino]-2-[[2,3-bis(oxidanyl)phenyl]carbonylamino]-3-oxidanylidene-propyl]amino]-2-[[2,3-bis(oxidanyl)phenyl]carbonylamino]-3-oxidanylidene-propyl]amino]-2-oxidanylidene-ethyl]-2,3-bis(oxidanyl)benzamide | | Authors: | Naismith, J.H, Moynie, L.M. | | Deposit date: | 2020-05-04 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.717 Å) | | Cite: | Hijacking of the Enterobactin Pathway by a Synthetic Catechol Vector Designed for Oxazolidinone Antibiotic Delivery in Pseudomonas aeruginosa.
Acs Infect Dis., 2022
|
|
6YYN
 
 | | Structure of Cathepsin S in complex with Compound 14 | | Descriptor: | CITRATE ANION, Cathepsin S, SULFATE ION, ... | | Authors: | Wagener, M, Schade, M, Merla, B, Hars, U, Kueckelhaus, S.Q. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Highly Selective Sub-Nanomolar Cathepsin S Inhibitors by Merging Fragment Binders with Nitrile Inhibitors.
J.Med.Chem., 63, 2020
|
|







