7XSB
 
 | |
8A99
 
 | | SARS Cov2 Spike in 1-up conformation complex with Fab47 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab47 (Heavy chain variable domain), ... | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2022-06-28 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Immunoglobulin germline gene polymorphisms influence the function of SARS-CoV-2 neutralizing antibodies.
Immunity, 56, 2023
|
|
8H3M
 
 | | Conformation 1 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy chain, Spike glycoprotein | | Authors: | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
8H3N
 
 | | Conformation 2 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy-chain, MO1 light chain, ... | | Authors: | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
8OT0
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with MTA and glycine | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Falke, S. | | Deposit date: | 2023-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
8OV1
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with ADP | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D. | | Deposit date: | 2023-04-25 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
8BSE
 
 | | CRYSTAL STRUCTURE OF SARS-COV-2 RECEPTOR BINDING DOMAIN (RBD) in complex with 1D1 Fab | | Descriptor: | 1D1 FAB HEAVY CHAIN, 1D1 FAB LIGHT CHAIN, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Welin, M, Kimbung, Y.R, Focht, D, Pisitkun, T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Efficacy of the combination of monoclonal antibodies against the SARS-CoV-2 Beta and Delta variants.
Plos One, 18, 2023
|
|
8BSF
 
 | | CRYSTAL STRUCTURE OF SARS-COV-2 RECEPTOR BINDING DOMAIN (RBD-beta variant) in complex with 3D2 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3D2 FAB HEAVY CHAIN, 3D2 FAB LIGHT CHAIN, ... | | Authors: | Welin, M, Kimbung, Y.R, Focht, D, Pisitkun, T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Efficacy of the combination of monoclonal antibodies against the SARS-CoV-2 Beta and Delta variants.
Plos One, 18, 2023
|
|
8CSJ
 
 | |
8FWN
 
 | | Crystal structure of SARS-CoV-2 papain-like protease C111S mutant | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Bezerra, E.H.S, Soprano, A.S, Tonoli, C.C.C, Prado, P.F.V, da Silva, J.C, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2023-01-23 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 papain-like protease C111S mutant
To Be Published
|
|
8HN6
 
 | | Crystal structure of monoclonal antibody complexed with SARS-CoV-2 RBD | | Descriptor: | Heavy chain of monoclonal antibody 3G10, Light chain of monoclonal antibody 3G10, Spike protein S1 | | Authors: | Qi, J, Chen, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Characterization of RBD-specific cross-neutralizing antibodies responses against SARS-CoV-2 variants from COVID-19 convalescents.
Front Immunol, 14, 2023
|
|
8HN7
 
 | | Crystal structure of monoclonal antibody complexed with SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of monoclonal antibody 3C11, Light chain of monoclonal antibody 3C11, ... | | Authors: | Qi, J, Chen, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Characterization of RBD-specific cross-neutralizing antibodies responses against SARS-CoV-2 variants from COVID-19 convalescents.
Front Immunol, 14, 2023
|
|
8IF2
 
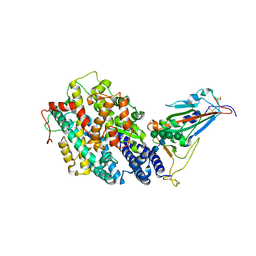 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BQ.1.1 variant spike protein in complex with its receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Kimura, K, Suzuki, T, Hashiguchi, T. | | Deposit date: | 2023-02-17 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Convergent evolution of SARS-CoV-2 Omicron subvariants leading to the emergence of BQ.1.1 variant.
Nat Commun, 14, 2023
|
|
7XSC
 
 | |
8CWI
 
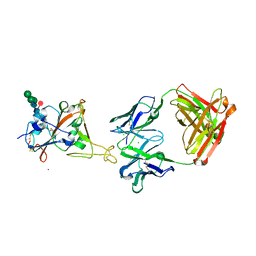 | | Fab arm of antibody 10G4 bound to CoV-2 receptor binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Heavy chain of Fab arm of antibody 10G4, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-24 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.873 Å) | | Cite: | Affinity maturation endows potent activity onto class 6 SARS-CoV-2 broadly neutralizing antibodies.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8CWK
 
 | |
8DNN
 
 | | Crystal structure of neutralizing antibody 80 in complex with SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 80 FAB HEAVY CHAIN, 80 FAB LIGHT CHAIN, ... | | Authors: | Muthuraman, K, Kucharska, I, Ivanochko, D, Julien, J.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A multi-specific, multi-affinity antibody platform neutralizes sarbecoviruses and confers protection against SARS-CoV-2 in vivo.
Sci Transl Med, 15, 2023
|
|
8GB5
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody 25F9 | | Descriptor: | 25F9 Heavy chain, 25F9 Light chain, BICINE, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-02-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Broadly neutralizing antibodies against sarbecoviruses generated by immunization of macaques with an AS03-adjuvanted COVID-19 vaccine.
Sci Transl Med, 15, 2023
|
|
8GB6
 
 | |
8GB7
 
 | |
8GB8
 
 | |
8ILC
 
 | | Crystal structure of Se-Met CoV-Y domain of Nsp3 in SARS-CoV-2 | | Descriptor: | CHLORIDE ION, Papain-like protease nsp3, SULFATE ION | | Authors: | Wang, K, Nan, J, Lei, J. | | Deposit date: | 2023-03-03 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The CoV-Y domain of SARS-CoV-2 Nsp3 interacts with BRAP to stimulate NF-kappa B signaling and induce host inflammatory responses.
Int.J.Biol.Macromol., 280, 2024
|
|
8IOS
 
 | | Structure of the SARS-CoV-2 XBB.1 spike glycoprotein (closed-1 state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8IOU
 
 | | Structure of SARS-CoV-2 XBB.1 spike glycoprotein in complex with ACE2 (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8IOV
 
 | | Structure of SARS-CoV-2 XBB.1 spike RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|








