8WE4
 
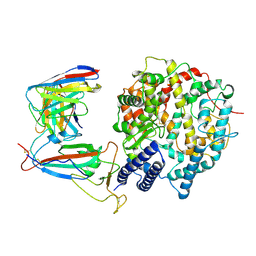 | | SARS-CoV-2 Omicron XBB.1.5 RBD complexed with human ACE2 and S304 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Xie, Y. | | Deposit date: | 2023-09-17 | | Release date: | 2024-07-24 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Key mechanistic features of the trade-off between antibody escape and host cell binding in the SARS-CoV-2 Omicron variant spike proteins.
Embo J., 43, 2024
|
|
8K9B
 
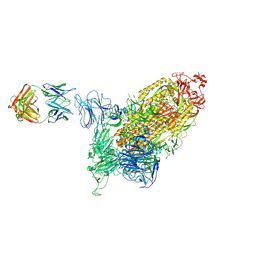 | | SARS-CoV-2 spike protein in complex with one S2H5 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of S2H5 Fab, Light chain of S2H5 Fab, ... | | Authors: | Hong, Q, Cong, Y. | | Deposit date: | 2023-07-31 | | Release date: | 2024-07-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular basis of SARS-CoV-2 immune evasion of NTD neutralizing antibodies.
Dian Zi Xian Wei Xue Bao, 2024
|
|
8K9J
 
 | |
8K9M
 
 | |
8TMY
 
 | |
8VCR
 
 | | SARS-CoV-2 Spike S2 bound to Fab 54043-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 54043-5 Fab Heavy Chain, ... | | Authors: | Johnson, N.V, McLellan, J.S. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Discovery and characterization of a pan-betacoronavirus S2-binding antibody.
Structure, 32, 2024
|
|
8YWW
 
 | | The structure of HKU1-B S protein with bsAb1 | | Descriptor: | H4B6 heavy chain, H4B6 light chain, Spike glycoprotein, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2024-04-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A novel bispecific antibody targeting two overlapping epitopes in RBD improves neutralizing potency and breadth against SARS-CoV-2.
Emerg Microbes Infect, 13, 2024
|
|
8YWX
 
 | |
9BRV
 
 | | SARS-CoV-2 Papain-like Protease (PLpro) with Fragment 5 | | Descriptor: | CHLORIDE ION, N-[2-(dimethylamino)ethyl]-N'-(3-methylphenyl)thiourea, Papain-like protease nsp3, ... | | Authors: | Amporndanai, K, Zhao, B, Fesik, S.W. | | Deposit date: | 2024-05-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fragment-Based Screen of SARS-CoV-2 Papain-like Protease (PL pro ).
Acs Med.Chem.Lett., 15, 2024
|
|
9BRW
 
 | | SARS-CoV-2 Papain-like Protease (PLpro) with Fragment 7 | | Descriptor: | CHLORIDE ION, N-(2-chlorophenyl)-1-methyl-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Papain-like protease nsp3, ... | | Authors: | Amporndanai, K, Zhao, B, Fesik, S.W. | | Deposit date: | 2024-05-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-Based Screen of SARS-CoV-2 Papain-like Protease (PL pro ).
Acs Med.Chem.Lett., 15, 2024
|
|
9BRX
 
 | | SARS-CoV-2 Papain-like Protease (PLpro) with Fragment 10 | | Descriptor: | (4R)-N-(2,4-dimethylphenyl)-7-methyl[1,2,4]triazolo[4,3-a]pyrimidin-5-amine, Papain-like protease nsp3, SULFATE ION, ... | | Authors: | Amporndanai, K, Zhao, B, Fesik, S.W. | | Deposit date: | 2024-05-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-Based Screen of SARS-CoV-2 Papain-like Protease (PL pro ).
Acs Med.Chem.Lett., 15, 2024
|
|
9CPO
 
 | |
9FX6
 
 | | Crystal structure of Cryo2RT SARS-CoV-2 main protease at 100K | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Huang, C.Y, Aumonier, S, Mac Sweeney, A, Olieric, V, Wang, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
9FX7
 
 | | Crystal structure of Cryo2RT SARS-CoV-2 main protease at 294K | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Huang, C.Y, Aumonier, S, Mac Sweeney, A, Olieric, V, Wang, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Cryo2RT: a high-throughput method for room-temperature macromolecular crystallography from cryo-cooled crystals.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
9IK2
 
 | | The co-crystal structure of SARS-CoV-2 Mpro in complex with compound H109 | | Descriptor: | 3C-like proteinase, tert-butyl N-[(2S)-1-[[(2S)-1-[[(2S)-1-azanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | Feng, Y, Zheng, W.Y, Han, P, Fu, L.F, Qi, J.X. | | Deposit date: | 2024-06-26 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided discovery of a small molecule inhibitor of SARS-CoV-2 main protease with potent in vitro and in vivo antiviral activities
To Be Published
|
|
8C9P
 
 | | Crystal structure of SARS-CoV-2 Mpro-E166V mutant, free enzyme | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | El Kilani, H, Ibrahim, M, Hilgenfeld, R. | | Deposit date: | 2023-01-23 | | Release date: | 2024-08-07 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 13b-K and Nirmatrelvir Resistance Mutations of SARS-CoV-2 Main Protease: Structural, Biochemical, and Biophysical Characterization of Free Enzymes and Inhibitor Complexes
Crystals, 15, 2025
|
|
8C9Q
 
 | | Crystal structure of SARS-CoV-2 Mpro-Q189K mutant in complex with 13b-K | | Descriptor: | Non-structural protein 11, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2023-01-23 | | Release date: | 2024-08-07 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | 13b-K and Nirmatrelvir Resistance Mutations of SARS-CoV-2 Main Protease: Structural, Biochemical, and Biophysical Characterization of Free Enzymes and Inhibitor Complexes
Crystals, 15, 2025
|
|
8C9U
 
 | | Crystal structure of SARS-CoV-2 Mpro-Q189K mutant in complex with nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, Non-structural protein 11 | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2023-01-23 | | Release date: | 2024-08-07 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 13b-K and Nirmatrelvir Resistance Mutations of SARS-CoV-2 Main Protease: Structural, Biochemical, and Biophysical Characterization of Free Enzymes and Inhibitor Complexes
Crystals, 15, 2025
|
|
8CA6
 
 | |
8CA8
 
 | | Crystal structure of SARS-CoV-2 Mpro-H172Y mutant, free enzyme | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | El Kilani, H, Ibrahim, M, Hilgenfeld, R. | | Deposit date: | 2023-01-24 | | Release date: | 2024-08-07 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 13b-K and Nirmatrelvir Resistance Mutations of SARS-CoV-2 Main Protease: Structural, Biochemical, and Biophysical Characterization of Free Enzymes and Inhibitor Complexes
Crystals, 15, 2025
|
|
8CAC
 
 | | Crystal structure of SARS-CoV-2 Mpro-H172Y mutant in complex with 13b-K | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Ibrahim, M, El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2023-01-24 | | Release date: | 2024-08-07 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | 13b-K and Nirmatrelvir Resistance Mutations of SARS-CoV-2 Main Protease: Structural, Biochemical, and Biophysical Characterization of Free Enzymes and Inhibitor Complexes
Crystals, 15, 2025
|
|
8CAJ
 
 | | Crystal structure of SARS-CoV-2 Mpro-E166V mutant in complex with 13b-K | | Descriptor: | Non-structural protein 11, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{S})-3-oxidanyl-4-oxidanylidene-1-[(3~{R})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2023-01-24 | | Release date: | 2024-08-07 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 13b-K and Nirmatrelvir Resistance Mutations of SARS-CoV-2 Main Protease: Structural, Biochemical, and Biophysical Characterization of Free Enzymes and Inhibitor Complexes
Crystals, 15, 2025
|
|
8SDH
 
 | |
8VQX
 
 | | Structure of SARS-CoV-2 main protease with potent peptide aldehyde inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dougan, D.R, Lane, W. | | Deposit date: | 2024-01-19 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Exploiting high-energy hydration sites for the discovery of potent peptide aldehyde inhibitors of the SARS-CoV-2 main protease with cellular antiviral activity.
Bioorg.Med.Chem., 103, 2024
|
|
9AUJ
 
 | | Structure of SARS-CoV-2 Mpro mutant (S144A) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.486 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|








