8IV5
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 1C4 (local refinement) | | Descriptor: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV8
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 3E2 and 1C4 (local refinement) | | Descriptor: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IVA
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs XMA01 and 3E2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8JOP
 
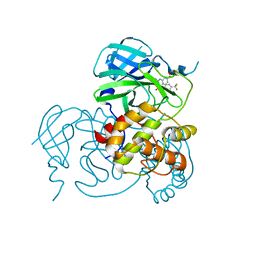 | | Crystal structure of the SARS-CoV-2 main protease in complex with 11a | | Descriptor: | 3C-like proteinase nsp5, methyl (6~{R})-5-ethanoyl-7-oxidanylidene-6-[4-(trifluoromethyl)phenyl]-8,9,10,11-tetrahydro-6~{H}-benzo[b][1,4]benzodiazepine-2-carboxylate | | Authors: | Zeng, R, Liu, Y.Z, Wang, F.L, Yang, S.Y, Lei, J. | | Deposit date: | 2023-06-08 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of benzodiazepine derivatives as a new class of covalent inhibitors of SARS-CoV-2 main protease.
Bioorg.Med.Chem.Lett., 92, 2023
|
|
8SK5
 
 | | Crystal structure of the SARS-CoV-2 neutralizing VHH 7A9 bound to the spike receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, anti-SARS-CoV-2 receptor binding domain VHH | | Authors: | Noland, C.L, Pande, K, Zhang, L, Zhou, H, Galli, J, Eddins, M, Gomez-Llorente, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Discovery and multimerization of cross-reactive single-domain antibodies against SARS-like viruses to enhance potency and address emerging SARS-CoV-2 variants.
Sci Rep, 13, 2023
|
|
8GF2
 
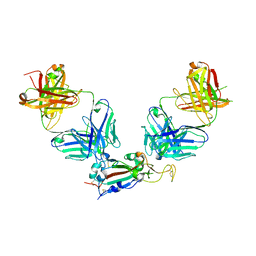 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies eCR3022.20 and CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 Fab heavy chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Broadening a SARS-CoV-1-neutralizing antibody for potent SARS-CoV-2 neutralization through directed evolution.
Sci.Signal., 16, 2023
|
|
8GW4
 
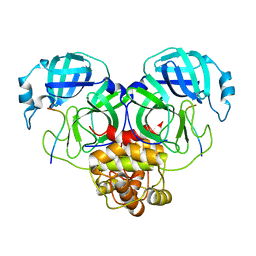 | | SARS-CoV-2 Mpro 1-302/C145A in complex with peptide 8-1 | | Descriptor: | Replicase polyprotein 1ab, peptide 8-1 | | Authors: | Liu, M, Huang, H. | | Deposit date: | 2022-09-16 | | Release date: | 2023-08-23 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The S1'-S3' Pocket of the SARS-CoV-2 Main Protease Is Critical for Substrate Selectivity and Can Be Targeted with Covalent Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8GWS
 
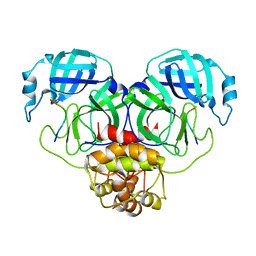 | | SARS-CoV-2 Mpro 1-302 c145a in complex with peptide 4 | | Descriptor: | Replicase polyprotein 1ab, VAL-LYS-LEU-GLN-ALA-ILE-PHE-ARG | | Authors: | Liu, M, Fu, Z, Huang, H. | | Deposit date: | 2022-09-17 | | Release date: | 2023-08-23 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The S1'-S3' Pocket of the SARS-CoV-2 Main Protease Is Critical for Substrate Selectivity and Can Be Targeted with Covalent Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8JPQ
 
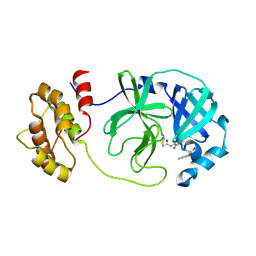 | | SARS-CoV-2 Mpro in complex with D-5-96 | | Descriptor: | 3C-like proteinase nsp5, ACE-ALA-ILE-V3E | | Authors: | Liu, M, Huang, H. | | Deposit date: | 2023-06-12 | | Release date: | 2023-08-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The S1'-S3' Pocket of the SARS-CoV-2 Main Protease Is Critical for Substrate Selectivity and Can Be Targeted with Covalent Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8BWT
 
 | |
8CYU
 
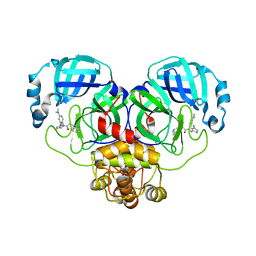 | | Crystal structure of SARS-CoV-2 Mpro with compound C5 | | Descriptor: | 3C-like proteinase, N-[(4-chlorothiophen-2-yl)methyl]-N-[4-(dimethylamino)phenyl]-2-(isoquinolin-4-yl)acetamide | | Authors: | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8CYZ
 
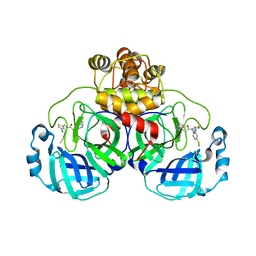 | | Crystal structure of SARS-CoV-2 Mpro with compound C4 | | Descriptor: | 3C-like proteinase, N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)-N-[4-(methylsulfanyl)phenyl]acetamide | | Authors: | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8CZ4
 
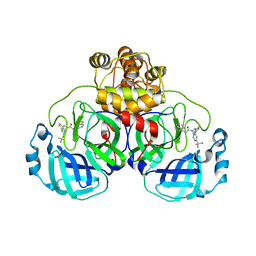 | | Crystal structure of SARS-CoV-2 Mpro with compound C3 | | Descriptor: | 3C-like proteinase, N-(4-tert-butylphenyl)-N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)acetamide | | Authors: | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8CZ7
 
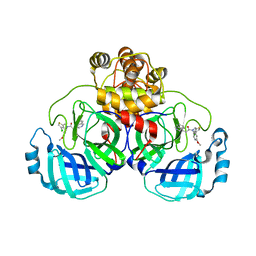 | | Crystal structure of SARS-CoV-2 Mpro with compound C2 | | Descriptor: | 3C-like proteinase, N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)-N-(4-methoxyphenyl)acetamide | | Authors: | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8FY6
 
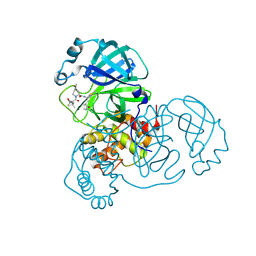 | | SARS-CoV-2 main protease in complex with covalent inhibitor | | Descriptor: | (1R,2S,5S)-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-N-{(2R)-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Fried, W, Chen, X.S. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Alkyne as a Latent Warhead to Covalently Target SARS-CoV-2 Main Protease.
J.Med.Chem., 66, 2023
|
|
8FY7
 
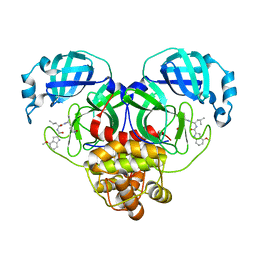 | | SARS-CoV-2 main protease in complex with covalent inhibitor | | Descriptor: | 3C-like proteinase nsp5, 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-1-[(3S)-2-oxopyrrolidin-3-yl]but-3-en-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide | | Authors: | Fried, W, Chen, X.S. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Alkyne as a Latent Warhead to Covalently Target SARS-CoV-2 Main Protease.
J.Med.Chem., 66, 2023
|
|
8GNH
 
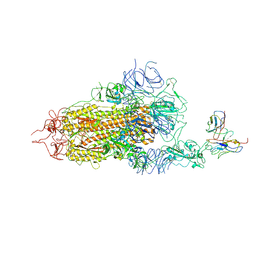 | | Complex structure of BD-218 and Spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BD-218, ... | | Authors: | Wang, B, Xu, H, Su, X.D. | | Deposit date: | 2022-08-23 | | Release date: | 2023-08-30 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Human antibody BD-218 has broad neutralizing activity against concerning variants of SARS-CoV-2.
Int.J.Biol.Macromol., 227, 2023
|
|
8GQT
 
 | |
8H7L
 
 | | Cryo-EM Structure of SARS-CoV-2 BA.2 Spike protein in complex with BA7535 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7535 fab heavt chain, ... | | Authors: | Liu, Z, Yan, A, Gao, Y. | | Deposit date: | 2022-10-20 | | Release date: | 2023-08-30 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Identification of a highly conserved neutralizing epitope within the RBD region of diverse SARS-CoV-2 variants.
Nat Commun, 15, 2024
|
|
8H7Z
 
 | |
8HFX
 
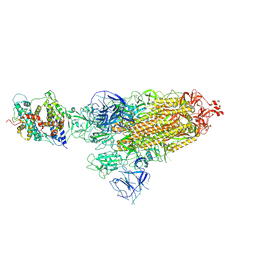 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
8HFY
 
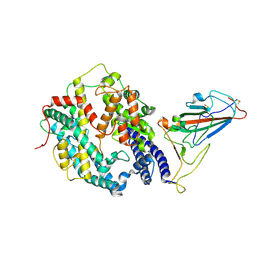 | | SARS-CoV-2 Omicron BA.1 spike protein receptor-binding domain in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
8HFZ
 
 | | Cryo-EM structure of SARS-CoV-2 prototype spike protein in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
8HG0
 
 | | Cryo-EM structure of SARS-CoV-2 prototype spike protein receptor-binding domain in complex with white-tailed deer ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Han, P, Meng, Y.M, Qi, J.X. | | Deposit date: | 2022-11-13 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural basis of white-tailed deer, Odocoileus virginianus , ACE2 recognizing all the SARS-CoV-2 variants of concern with high affinity.
J.Virol., 97, 2023
|
|
8HTV
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3a | | Descriptor: | 1-(5,6-dihydrobenzo[b][1]benzazepin-11-yl)-2-sulfanyl-ethanone, 3C-like proteinase | | Authors: | Su, H.X, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Discovery and Mechanism Study of SARS-CoV-2 3C-like Protease Inhibitors with a New Reactive Group.
J.Med.Chem., 66, 2023
|
|








