7T2T
 
 | |
7T2U
 
 | |
7T2V
 
 | |
7XXL
 
 | | RBD in complex with Fab14 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab14 heavy chain, Fab14 light chain, ... | | 著者 | Lin, J.Q, Tan, Y.J.E, Wu, B, Lescar, J. | | 登録日 | 2022-05-30 | | 公開日 | 2022-09-14 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (7.3 Å) | | 主引用文献 | Engineering SARS-CoV-2 specific cocktail antibodies into a bispecific format improves neutralizing potency and breadth.
Nat Commun, 13, 2022
|
|
8E64
 
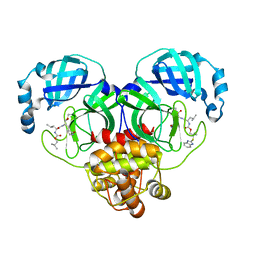 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a benzimidazole dimethyl sulfane inhibitor | | 分子名称: | (1S,2S)-2-{[N-({2-[(1H-benzimidazol-2-yl)sulfanyl]-2-methylpropoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1~{R},2~{S})-2-[[(2~{S})-2-[[2-(1~{H}-benzimidazol-2-ylsulfanyl)-2-methyl-propoxy]carbonylamino]-4-methyl-pentanoyl]amino]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propane-1-sulfonic acid;molecular oxygen, 3C-like proteinase | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Madden, T.K, Groutas, W.C. | | 登録日 | 2022-08-22 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8E65
 
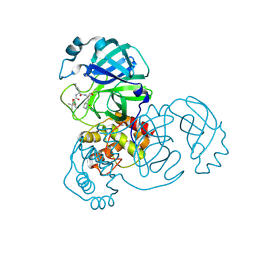 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a p-chlorodimethyl oxybenzene inhibitor | | 分子名称: | (1S,2S)-2-[(N-{[2-(4-chlorophenoxy)-2-methylpropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Miller, M.J, Groutas, W.C. | | 登録日 | 2022-08-22 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8E68
 
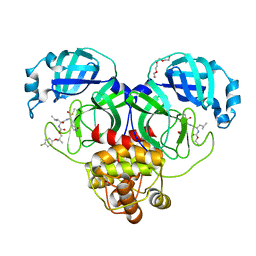 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a p-fluorodimethyl oxybenzene inhibitor | | 分子名称: | (1S,2S)-2-[(N-{[2-(4-fluorophenoxy)-2-methylpropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, N~2~-{[2-(4-fluorophenoxy)-2-methylpropoxy]carbonyl}-N-{(1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}-L-leucinamide, ... | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Miller, M.J, Groutas, W.C. | | 登録日 | 2022-08-22 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
7SA2
 
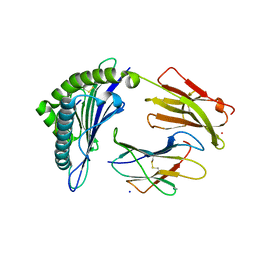 | |
7XCO
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with S309 fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 Fab heavy chain, ... | | 著者 | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | 登録日 | 2022-03-24 | | 公開日 | 2022-09-21 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7Y9Z
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with human ACE2 ectodomain (one-RBD-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Gao, G.F, Qi, J.X, Liu, S, Zhao, Z.N. | | 登録日 | 2022-06-26 | | 公開日 | 2022-09-21 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7YA0
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (S-6P-RRAR) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2022-06-26 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
8DOX
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-245 | | 分子名称: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | 登録日 | 2022-07-14 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Identification of SARS-CoV-2 M pro inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2.
Nat Commun, 14, 2023
|
|
8DRR
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp4-nsp5 (C4) cut site sequence | | 分子名称: | 3C-like proteinase nsp5, SODIUM ION | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRS
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp6-nsp7 (C6) cut site sequence | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRT
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp6-nsp7 (C6) cut site sequence (form 2) | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRU
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp7-nsp8 (C7) cut site sequence | | 分子名称: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp7-nsp8 (C7) cut site, PENTAETHYLENE GLYCOL, ... | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRV
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp8-nsp9 (C8) cut site sequence | | 分子名称: | Fusion protein of 3C-like proteinase nsp5 and nsp8-nsp9 (C8) cut site, PENTAETHYLENE GLYCOL | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRW
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp9-nsp10 (C9) cut site sequence | | 分子名称: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp9-nsp10 (C9) cut site, PENTAETHYLENE GLYCOL, ... | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRX
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp10-nsp11 (C10) cut site sequence (form 2) | | 分子名称: | Fusion protein of 3C-like proteinase nsp5 and nsp10-nsp11 (C10) cut site, SODIUM ION | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRY
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp12-nsp13 (C12) cut site sequence | | 分子名称: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp12-nsp13 (C12) cut site | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8E5Z
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a dimethyl sulfonyl benzene inhibitor | | 分子名称: | (1R,2S)-2-[(N-{[2-(benzenesulfonyl)-2-methylpropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-[(N-{[2-(benzenesulfonyl)-2-methylpropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Miller, M.J, Groutas, W.C. | | 登録日 | 2022-08-22 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8E69
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a m-fluorodimethyl oxybenzene inhibitor | | 分子名称: | (1R,2S)-2-[(N-{[2-(3-fluorophenoxy)-2-methylpropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Miller, M.J, Groutas, W.C. | | 登録日 | 2022-08-22 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8E6A
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a p-chlorophenylethanol based inhibitor | | 分子名称: | (1R,2S)-2-[(N-{[(2S)-2-(3-chlorophenyl)-2-hydroxypropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl]propane-1-sulfonic acid, (1S,2S)-2-[(N-{[(2R)-2-(3-chlorophenyl)-2-hydroxypropoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | 著者 | Lovell, S, Liu, L, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2022-08-22 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
7WRJ
 
 | |
7XWA
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BA.4/5 variant spike protein in complex with its receptor ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Suzuki, T, Kimura, K, Hashiguchi, T. | | 登録日 | 2022-05-26 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.36 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5.
Cell, 185, 2022
|
|







