7ULT
 
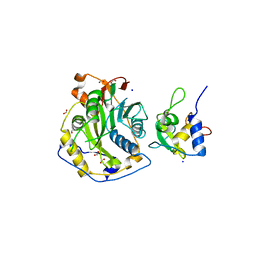 | | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer Apo-Form. | | Descriptor: | 2'-O-methyltransferase, FORMIC ACID, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-04-05 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer Apo-Form.
To Be Published
|
|
7WED
 
 | |
7WLC
 
 | |
7WO1
 
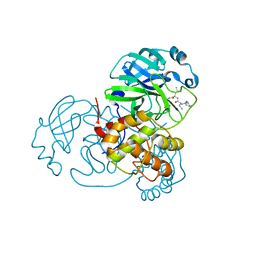 | | Discovery of SARS-CoV-2 3CLpro peptidomimetic inhibitors through H41-specific protein-ligand interactions | | Descriptor: | 3C-like proteinase, N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]cyclohexanecarboxamide | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-20 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
7WO3
 
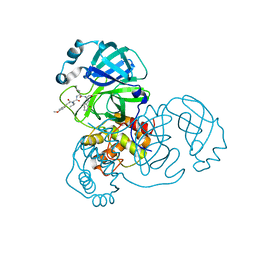 | | SARS-CoV-2 3CLpro | | Descriptor: | (2S)-2-[[(2S)-2-[[(E)-3-(4-methoxyphenyl)prop-2-enoyl]amino]-3-methyl-butanoyl]amino]-4-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]pentanamide, 3C-like proteinase | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-20 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
7WOF
 
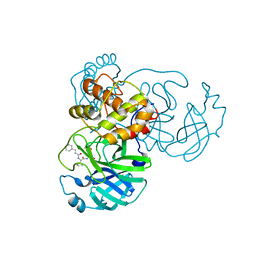 | | SARS-CoV-2 3CLpro | | Descriptor: | (2S,3S)-3-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-2-[[(E)-3-phenylprop-2-enoyl]amino]pentanamide, 3C-like proteinase | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-21 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
7WOH
 
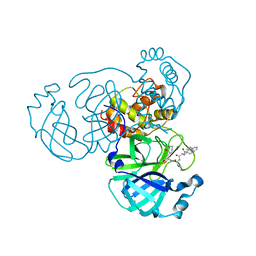 | | SARS-CoV-2 3CLpro | | Descriptor: | (2S)-4-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-2-[[(2S)-3-phenyl-2-[[(E)-3-phenylprop-2-enoyl]amino]propanoyl]amino]pentanamide, 3C-like proteinase | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-21 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
7X7D
 
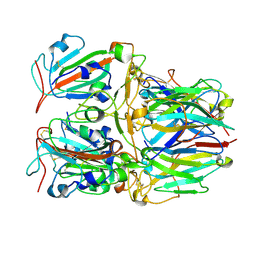 | | SARS-CoV-2 Delta RBD and Nb22 | | Descriptor: | Nb22, Spike protein S1 | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-03-09 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Short-Term Instantaneous Prophylaxis and Efficient Treatment Against SARS-CoV-2 in hACE2 Mice Conferred by an Intranasal Nanobody (Nb22).
Front Immunol, 13, 2022
|
|
7Z0X
 
 | | THSC20.HVTR26 Fab bound to SARS-CoV-2 Receptor Binding Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Spike protein S1, ... | | Authors: | Wibmer, C.K. | | Deposit date: | 2022-02-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A combination of potently neutralizing monoclonal antibodies isolated from an Indian convalescent donor protects against the SARS-CoV-2 Delta variant.
Plos Pathog., 18, 2022
|
|
7Z0Y
 
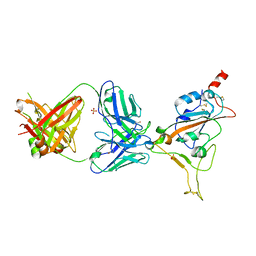 | | THSC20.HVTR04 Fab bound to SARS-CoV-2 Receptor Binding Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Spike protein S1, ... | | Authors: | Wibmer, C.K. | | Deposit date: | 2022-02-23 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A combination of potently neutralizing monoclonal antibodies isolated from an Indian convalescent donor protects against the SARS-CoV-2 Delta variant.
Plos Pathog., 18, 2022
|
|
7QCR
 
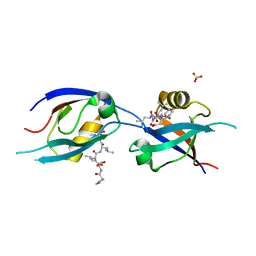 | | MLLT4/Afadin PDZ domain in complex with the C-terminal peptide from protein E of SARS-CoV-2 | | Descriptor: | Afadin, Envelope small membrane protein, SULFATE ION | | Authors: | Zhu, Y, Alvarez, F, Haouz, A, Mechaly, A, Caillet-Saguy, C. | | Deposit date: | 2021-11-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Interactions of Severe Acute Respiratory Syndrome Coronavirus 2 Protein E With Cell Junctions and Polarity PSD-95/Dlg/ZO-1-Containing Proteins.
Front Microbiol, 13, 2022
|
|
7QCS
 
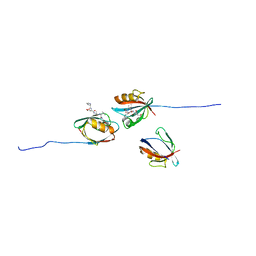 | | PALS1/MPP5 PDZ domain in complex with SARS-CoV-2_E PBM peptide | | Descriptor: | Envelope small membrane protein, Protein PALS1 | | Authors: | Zhu, Y, Alvarez, F, Haouz, A, Mechaly, A, Caillet-Saguy, C. | | Deposit date: | 2021-11-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Interactions of Severe Acute Respiratory Syndrome Coronavirus 2 Protein E With Cell Junctions and Polarity PSD-95/Dlg/ZO-1-Containing Proteins.
Front Microbiol, 13, 2022
|
|
7QCT
 
 | | PDZ2 of LNX2 with SARS-CoV-2_E PBM complex | | Descriptor: | Envelope small membrane protein, Ligand of Numb protein X 2 | | Authors: | Zhu, Y, Alvarez, F, Haouz, A, Mechaly, A, Caillet-Saguy, C. | | Deposit date: | 2021-11-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.197 Å) | | Cite: | Interactions of Severe Acute Respiratory Syndrome Coronavirus 2 Protein E With Cell Junctions and Polarity PSD-95/Dlg/ZO-1-Containing Proteins.
Front Microbiol, 13, 2022
|
|
7R40
 
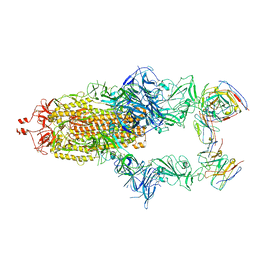 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the 87G7 antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 87G7 heavy chain variable region, ... | | Authors: | Hurdiss, D.L. | | Deposit date: | 2022-02-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | An ACE2-blocking antibody confers broad neutralization and protection against Omicron and other SARS-CoV-2 variants of concern.
Sci Immunol, 7, 2022
|
|
7RQ6
 
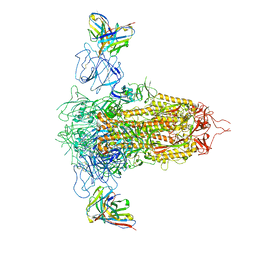 | | Cryo-EM structure of SARS-CoV-2 spike in complex with non-neutralizing NTD-directed CV3-13 Fab isolated from convalescent individual | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CV3-13 Fab heavy chain, ... | | Authors: | Chen, Y, Pozharski, E, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2021-08-05 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | A Fc-enhanced NTD-binding non-neutralizing antibody delays virus spread and synergizes with a nAb to protect mice from lethal SARS-CoV-2 infection.
Cell Rep, 38, 2022
|
|
7TPR
 
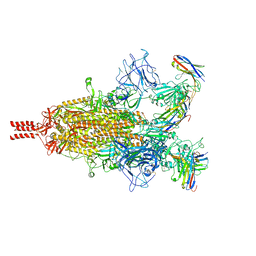 | | Camel nanobodies 7A3 and 8A2 broadly neutralize SARS-CoV-2 variants | | Descriptor: | Nanobody 7A3, Nanobody 8A2, Spike glycoprotein | | Authors: | Butay, K.J, Zhu, J, Dandey, V.P, Hong, J, Kwon, H.J, Chen, C.Z, Duan, Z, Li, D, Ren, H, Liang, T, Martin, N, Esposito, D, Ortega-Rodriguez, U, Xu, M, Xie, H, Ho, M, Cachau, R, Borgnia, M.J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-20 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Camel nanobodies broadly neutralize SARS-CoV-2 variants
bioRxiv, 2021
|
|
7UB0
 
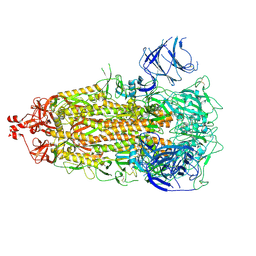 | |
7UB5
 
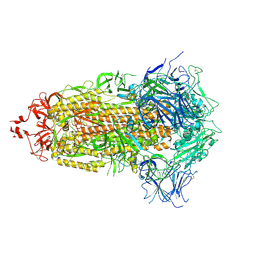 | |
7UB6
 
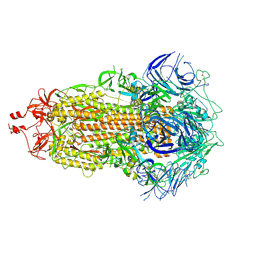 | |
7X7E
 
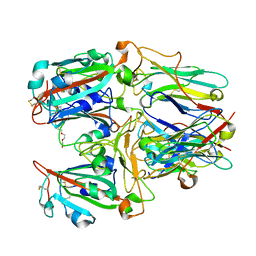 | | SARS-CoV-2 RBD and Nb22 | | Descriptor: | Nb22, Spike protein S1, TETRAETHYLENE GLYCOL | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-03-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Short-Term Instantaneous Prophylaxis and Efficient Treatment Against SARS-CoV-2 in hACE2 Mice Conferred by an Intranasal Nanobody (Nb22).
Front Immunol, 13, 2022
|
|
7FJN
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with two T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7FJS
 
 | | Crystal structure of T6 Fab bound to theSARS-CoV-2 RBD of B.1.351 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7PBE
 
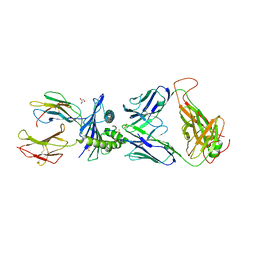 | | Emergence of immune escape at dominant SARS-CoV-2 killer T-cell epitope | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, Human T-cell Receptor YLQ36, ... | | Authors: | Rizkallah, P.J, Sewell, A.K, Wall, A, Fuller, A. | | Deposit date: | 2021-08-02 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Emergence of immune escape at dominant SARS-CoV-2 killer T cell epitope.
Cell, 185, 2022
|
|
7QL8
 
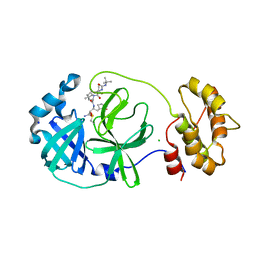 | | SARS-COV2 Main Protease in complex with inhibitor MG78 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2021-12-19 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | From Repurposing to Redesign: Optimization of Boceprevir to Highly Potent Inhibitors of the SARS-CoV-2 Main Protease.
Molecules, 27, 2022
|
|
7SD5
 
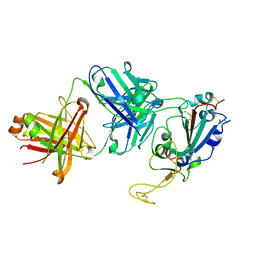 | | Crystallographic structure of neutralizing antibody 10-40 in complex with SARS-CoV-2 spike receptor binding domain | | Descriptor: | 10-40 Heavy chain, 10-40 Light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Reddem, E.R, Casner, R.G, Shapiro, L. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | An antibody class with a common CDRH3 motif broadly neutralizes sarbecoviruses.
Sci Transl Med, 14, 2022
|
|








