+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2w3m | ||||||
|---|---|---|---|---|---|---|---|
| Title | HUMAN DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADPH AND FOLATE | ||||||
 Components Components | DIHYDROFOLATE REDUCTASE | ||||||
 Keywords Keywords | OXIDOREDUCTASE / NONCLASSICAL ANTIFOLATES / ONE-CARBON METABOLISM / LIPOPHILIC ANTIFOLATES / NADP / REDUCTASE | ||||||
| Function / homology |  Function and homology information Function and homology informationregulation of removal of superoxide radicals / tetrahydrobiopterin biosynthetic process / Metabolism of folate and pterines / tetrahydrofolate metabolic process / response to methotrexate / sequence-specific mRNA binding / folic acid binding / axon regeneration / dihydrofolate metabolic process / G1/S-Specific Transcription ...regulation of removal of superoxide radicals / tetrahydrobiopterin biosynthetic process / Metabolism of folate and pterines / tetrahydrofolate metabolic process / response to methotrexate / sequence-specific mRNA binding / folic acid binding / axon regeneration / dihydrofolate metabolic process / G1/S-Specific Transcription / dihydrofolate reductase / dihydrofolate reductase activity / folic acid metabolic process / tetrahydrofolate biosynthetic process / NADPH binding / Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation / one-carbon metabolic process / mRNA regulatory element binding translation repressor activity / NADP binding / negative regulation of translation / mRNA binding / mitochondrion / cytosol Similarity search - Function | ||||||
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.6 Å MOLECULAR REPLACEMENT / Resolution: 1.6 Å | ||||||
 Authors Authors | Leung, A.K.W. / Reynolds, R.C. / Borhani, D.W. | ||||||
 Citation Citation |  Journal: To be Published Journal: To be PublishedTitle: Structural Basis for Selective Inhibition of Mycobacterium Avium Dihydrofolate Reductase by a Lipophilic Antifolate Authors: Leung, A.K.W. / Ross, L.J. / Zywno-Van Ginkel, S. / Reynolds, R.C. / Seitz, L.E. / Pathak, V. / Barrow, W.W. / White, E.L. / Suling, W.J. / Piper, J.R. / Borhani, D.W. | ||||||
| History |
| ||||||
| Remark 700 | SHEET THE SHEET STRUCTURE OF THIS MOLECULE IS BIFURCATED. IN ORDER TO REPRESENT THIS FEATURE IN ... SHEET THE SHEET STRUCTURE OF THIS MOLECULE IS BIFURCATED. IN ORDER TO REPRESENT THIS FEATURE IN THE SHEET RECORDS BELOW, TWO SHEETS ARE DEFINED. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2w3m.cif.gz 2w3m.cif.gz | 193.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2w3m.ent.gz pdb2w3m.ent.gz | 154.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2w3m.json.gz 2w3m.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/w3/2w3m https://data.pdbj.org/pub/pdb/validation_reports/w3/2w3m ftp://data.pdbj.org/pub/pdb/validation_reports/w3/2w3m ftp://data.pdbj.org/pub/pdb/validation_reports/w3/2w3m | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2w3aC  2w3bC  2w3vC  2w3wC  1drfS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 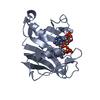
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
| ||||||||
| Noncrystallographic symmetry (NCS) | NCS oper: (Code: given Matrix: (-0.066385, 0.997007, -0.039625), Vector: |
- Components
Components
| #1: Protein | Mass: 21480.723 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Plasmid: PDFR / Production host: HOMO SAPIENS (human) / Plasmid: PDFR / Production host:  #2: Chemical | #3: Chemical | #4: Water | ChemComp-HOH / | Nonpolymer details | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE (NDP): THE NADPH MAY BE A MIXTURE WITH ...NADPH DIHYDRO-NICOTINAMI | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.35 Å3/Da / Density % sol: 47.72 % / Description: NONE |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 8.75 Details: HUMAN DHFR/FOLATE COMPLEX WAS MIXED WITH NADPH (2 MM FINAL). CRYSTALS WERE GROWN BY HANGING DROP VAPOR DIFFUSION AT 277 K BY MIXING EQUAL VOLUMES OF PROTEIN/NADPH/FOLATE WITH RESERVOIR (24% ...Details: HUMAN DHFR/FOLATE COMPLEX WAS MIXED WITH NADPH (2 MM FINAL). CRYSTALS WERE GROWN BY HANGING DROP VAPOR DIFFUSION AT 277 K BY MIXING EQUAL VOLUMES OF PROTEIN/NADPH/FOLATE WITH RESERVOIR (24% PEG 4000, 200 MM LI2SO4, 100 MM TRIS.HCL, PH 8.75). TRUNCATED TRIANGULAR CRYSTALS APPEARED SLOWLY, IN ABOUT A MONTH. THE CRYSTAL WAS CRYOPROTECTED WITH 15% GLYCEROL AND FLASH-COOLED IN LIQUID N2. |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  CHESS CHESS  / Beamline: A1 / Wavelength: 0.914 / Beamline: A1 / Wavelength: 0.914 |
| Detector | Type: ADSC QUANTUM-1 / Detector: CCD / Date: Jun 28, 1996 / Details: MIRRORS |
| Radiation | Monochromator: SI / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.914 Å / Relative weight: 1 |
| Reflection | Resolution: 1.6→10.7 Å / Num. obs: 52618 / % possible obs: 98.4 % / Observed criterion σ(I): -10 / Redundancy: 5.6 % / Biso Wilson estimate: 19.9 Å2 / Rmerge(I) obs: 0.05 / Net I/σ(I): 4.5 |
| Reflection shell | Resolution: 1.6→1.69 Å / Redundancy: 3.1 % / Rmerge(I) obs: 0.35 / Mean I/σ(I) obs: 1.8 / % possible all: 93.6 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1DRF Resolution: 1.6→10.7 Å / Cor.coef. Fo:Fc: 0.963 / Cor.coef. Fo:Fc free: 0.949 / SU B: 3.7 / SU ML: 0.06 / Cross valid method: THROUGHOUT / ESU R: 0.134 / ESU R Free: 0.094 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. THE OXIDATION STATE OF THE FOLATE LIGAND IS UNCLEAR FROM THE EXPERIMENTAL ELECTRON DENSITY MAPS. GIVEN THAT THE CRYSTAL WAS PREPARED WITH ...Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. THE OXIDATION STATE OF THE FOLATE LIGAND IS UNCLEAR FROM THE EXPERIMENTAL ELECTRON DENSITY MAPS. GIVEN THAT THE CRYSTAL WAS PREPARED WITH NADPH PRESENT, THE LIGAND MAY BE A MIXTURE OF FOLATE, 7,8-DIHYDROFOLATE, AND 5,6,7,8- TETRAHYDROFOLATE. IN ADDITION, THE OCCUPANCY ASSIGNED TO FOLATE, 0.8, WAS CHOSEN SO THAT THE AVERAGE TEMPERATURE FACTOR FOR THE PTERIDINE RING WAS SIMILAR TO SURROUNDING RESIDUES IN THE ACTIVE SITE. ACCORDINGLY, THE NADPH MAY BE A MIXTURE WITH THE OXIDIZED CO-FACTOR, NADP(+).
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: BABINET MODEL WITH MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 14.09 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.6→10.7 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller





































 PDBj
PDBj