[English] 日本語
 Yorodumi
Yorodumi- PDB-1cwa: X-RAY STRUCTURE OF A MONOMERIC CYCLOPHILIN A-CYCLOSPORIN A CRYSTA... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1cwa | ||||||
|---|---|---|---|---|---|---|---|
| Title | X-RAY STRUCTURE OF A MONOMERIC CYCLOPHILIN A-CYCLOSPORIN A CRYSTAL COMPLEX AT 2.1 ANGSTROMS RESOLUTION | ||||||
 Components Components |
| ||||||
 Keywords Keywords | ISOMERASE/IMMUNOSUPPRESSANT / ISOMERASE-IMMUNOSUPPRESSANT COMPLEX / CYCLOPHILIN-CYCLOSPORIN COMPLEX / CYCLOSPORIN A / IMMUNOSUPPRESSANT / CYCLOPHILIN | ||||||
| Function / homology |  Function and homology information Function and homology information: / negative regulation of protein K48-linked ubiquitination / regulation of apoptotic signaling pathway / cell adhesion molecule production / negative regulation of viral life cycle / lipid droplet organization / heparan sulfate binding / regulation of viral genome replication / virion binding / leukocyte chemotaxis ...: / negative regulation of protein K48-linked ubiquitination / regulation of apoptotic signaling pathway / cell adhesion molecule production / negative regulation of viral life cycle / lipid droplet organization / heparan sulfate binding / regulation of viral genome replication / virion binding / leukocyte chemotaxis / negative regulation of stress-activated MAPK cascade / endothelial cell activation / Basigin interactions / protein peptidyl-prolyl isomerization / cyclosporin A binding / Minus-strand DNA synthesis / Plus-strand DNA synthesis / Uncoating of the HIV Virion / Early Phase of HIV Life Cycle / Integration of provirus / APOBEC3G mediated resistance to HIV-1 infection / viral release from host cell / negative regulation of protein phosphorylation / Calcineurin activates NFAT / Binding and entry of HIV virion / positive regulation of viral genome replication / negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway / activation of protein kinase B activity / neutrophil chemotaxis / Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation / peptidyl-prolyl cis-trans isomerase activity / negative regulation of protein kinase activity / RNA polymerase II CTD heptapeptide repeat P3 isomerase activity / RNA polymerase II CTD heptapeptide repeat P6 isomerase activity / positive regulation of protein secretion / peptidylprolyl isomerase / Assembly Of The HIV Virion / Budding and maturation of HIV virion / platelet activation / platelet aggregation / neuron differentiation / positive regulation of NF-kappaB transcription factor activity / SARS-CoV-1 activates/modulates innate immune responses / integrin binding / unfolded protein binding / Platelet degranulation / protein folding / positive regulation of protein phosphorylation / cellular response to oxidative stress / secretory granule lumen / vesicle / ficolin-1-rich granule lumen / positive regulation of MAPK cascade / focal adhesion / intracellular membrane-bounded organelle / apoptotic process / Neutrophil degranulation / protein-containing complex / extracellular space / RNA binding / extracellular exosome / extracellular region / nucleus / membrane / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) TOLYPOCLADIUM INFLATUM (fungus) TOLYPOCLADIUM INFLATUM (fungus) | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 2.1 Å X-RAY DIFFRACTION / Resolution: 2.1 Å | ||||||
 Authors Authors | Mikol, V. / Kallen, J. / Walkinshaw, M.D. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1993 Journal: J.Mol.Biol. / Year: 1993Title: X-Ray Structure of a Monomeric Cyclophilin A-Cyclosporin a Crystal Complex at 2.1 A Resolution. Authors: Mikol, V. / Kallen, J. / Pflugl, G. / Walkinshaw, M.D. #1: Journal: Acta Crystallogr.,Sect.D / Year: 1994 Title: Crystallization of the Complex between Cyclophilin a and Cyclosporin Derivatives: The Use of Cross- Seeding. Authors: Mikol, V. / Duc, D. #2:  Journal: Protein Eng. / Year: 1994 Journal: Protein Eng. / Year: 1994Title: The X-Ray Structure of (Mebm2T)1-Cyclosporin Complexed with Cyclophilin a Provides an Explanation for its Anomalously High Immunosuppressive Activity. Authors: Mikol, V. / Kallen, J. / Walkinshaw, M.D. #3:  Journal: J.Med.Chem. / Year: 1994 Journal: J.Med.Chem. / Year: 1994Title: Improved Binding Affinity for Cyclophilin a by a Cyclosporin Derivative Singly Modified at its Effector Domain. Authors: Papageorgiou, C. / Florineth, A. / Mikol, V. #4: Journal: Nature / Year: 1993 Title: X-Ray Structure of a Decameric Cyclophilin- Cyclosporin Crystal Complex. Authors: Pflugl, G. / Kallen, J. / Schirmer, T. / Jansonius, J.N. / Zurini, M.G. / Walkinshaw, M.D. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1cwa.cif.gz 1cwa.cif.gz | 51.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1cwa.ent.gz pdb1cwa.ent.gz | 36.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1cwa.json.gz 1cwa.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cw/1cwa https://data.pdbj.org/pub/pdb/validation_reports/cw/1cwa ftp://data.pdbj.org/pub/pdb/validation_reports/cw/1cwa ftp://data.pdbj.org/pub/pdb/validation_reports/cw/1cwa | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 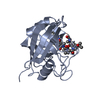
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Atom site foot note | 1: VAL C 5 - LEU C 6 OMEGA = 0.00 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION |
- Components
Components
| #1: Protein | Mass: 18036.504 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  HOMO SAPIENS (human) / Gene: CYCLOPHILIN / Gene (production host): CYCLOPHILIN / Production host: HOMO SAPIENS (human) / Gene: CYCLOPHILIN / Gene (production host): CYCLOPHILIN / Production host:  References: UniProt: P05092, UniProt: P62937*PLUS, peptidylprolyl isomerase |
|---|---|
| #2: Protein/peptide | 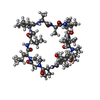  Type: Cyclic peptide / Class: Immunosuppressant / Mass: 1220.625 Da / Num. of mol.: 1 / Source method: obtained synthetically Type: Cyclic peptide / Class: Immunosuppressant / Mass: 1220.625 Da / Num. of mol.: 1 / Source method: obtained syntheticallyDetails: CYCLOSPORIN IS A CYCLIC UNDECAPEPTIDE. CYCLIZATION IS ACHIEVED BY LINKING THE N- AND THE C- TERMINI. Source: (synth.)  TOLYPOCLADIUM INFLATUM (fungus) / References: NOR: NOR00033, Cyclosporin A TOLYPOCLADIUM INFLATUM (fungus) / References: NOR: NOR00033, Cyclosporin A |
| #3: Water | ChemComp-HOH / |
| Compound details | CYCLOSPORI |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.07 Å3/Da / Density % sol: 40.61 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal | *PLUS Density % sol: 42 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 22 ℃ / pH: 8 / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Radiation | Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| Reflection | Resolution: 2.1→15 Å / Num. obs: 8732 / % possible obs: 88.9 % / Observed criterion σ(I): 2 |
| Reflection | *PLUS Num. measured all: 21441 / Rmerge(I) obs: 0.074 |
| Reflection shell | *PLUS Highest resolution: 2.1 Å / Lowest resolution: 2.24 Å / % possible obs: 55.8 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 2.1→8 Å / Rfactor Rwork: 0.167 / Rfactor obs: 0.167 / σ(F): 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.1→8 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller









































 PDBj
PDBj












