[English] 日本語
 Yorodumi
Yorodumi- PDB-2rmc: Crystal structure of murine cyclophilin C complexed with immunosu... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2rmc | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of murine cyclophilin C complexed with immunosuppressive drug cyclosporin A | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | ISOMERASE/IMMUNOSUPPRESSANT / ISOMERASE-IMMUNOSUPPRESSANT COMPLEX / CYCLOPHILIN-CYCLOSPORIN COMPLEX / CYCLOSPORIN A / IMMUNOSUPPRESSANT / CYCLOPHILIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpeptidylprolyl isomerase / peptidyl-prolyl cis-trans isomerase activity / protein folding / cytoplasm Similarity search - Function | |||||||||
| Biological species |   TOLYPOCLADIUM INFLATUM (fungus) TOLYPOCLADIUM INFLATUM (fungus) | |||||||||
| Method |  X-RAY DIFFRACTION / Resolution: 1.64 Å X-RAY DIFFRACTION / Resolution: 1.64 Å | |||||||||
 Authors Authors | Ke, H. / Zhao, Y. / Luo, F. / Weissman, I. / Friedman, J. | |||||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 1993 Journal: Proc.Natl.Acad.Sci.USA / Year: 1993Title: Crystal Structure of Murine Cyclophilin C Complexed with Immunosuppressive Drug Cyclosporin A Authors: Ke, H. / Zhao, Y. / Luo, F. / Weissman, I. / Friedman, J. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2rmc.cif.gz 2rmc.cif.gz | 205.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2rmc.ent.gz pdb2rmc.ent.gz | 168.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2rmc.json.gz 2rmc.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/rm/2rmc https://data.pdbj.org/pub/pdb/validation_reports/rm/2rmc ftp://data.pdbj.org/pub/pdb/validation_reports/rm/2rmc ftp://data.pdbj.org/pub/pdb/validation_reports/rm/2rmc | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
| ||||||||
| Atom site foot note | 1: RESIDUE ALA 8 IN CHAINS B, D, F, AND H IS A D-ALANINE. |
- Components
Components
| #1: Protein | Mass: 19941.682 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  #2: Protein/peptide | 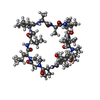  Type: Cyclic peptide / Class: Immunosuppressant / Mass: 1220.625 Da / Num. of mol.: 4 / Source method: obtained synthetically Type: Cyclic peptide / Class: Immunosuppressant / Mass: 1220.625 Da / Num. of mol.: 4 / Source method: obtained syntheticallyDetails: CYCLOSPORIN IS A CYCLIC UNDECAPEPTIDE. CYCLIZATION IS ACHIEVED BY LINKING THE N- AND THE C- TERMINI. Source: (synth.)  TOLYPOCLADIUM INFLATUM (fungus) / References: NOR: NOR00033, Cyclosporin A TOLYPOCLADIUM INFLATUM (fungus) / References: NOR: NOR00033, Cyclosporin A#3: Water | ChemComp-HOH / | Compound details | CYCLOSPORI | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.58 Å3/Da / Density % sol: 52.31 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS Temperature: 4 ℃ / pH: 6 / Method: microdialysis | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| Reflection | *PLUS Highest resolution: 1.64 Å / Num. obs: 73850 / Num. measured all: 139812 / Rmerge(I) obs: 0.04 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 1.64→6 Å Details: THE SPACE GROUP IS P21 WITH THE ORIGIN AT 1/4 OF X SO THAT THE SYMMETRY IS (X,Y,Z) AND (1/2-X, 1/2+Y,-X).
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.64→6 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Classification: refinement X-PLOR / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.197 / Rfactor Rwork: 0.197 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS Type: x_angle_d / Dev ideal: 2.68 |
 Movie
Movie Controller
Controller








































 PDBj
PDBj


