-Search query
-Search result
Showing all 46 items for (author: yang & zq)

EMDB-66856: 
Omicron-specific ultra-potent SARS-CoV-2 neutralizing antibodies targeting the N1/N2 loop of Spike N-terminal domain
Method: single particle / : Li ZQ, Niu X

EMDB-71559: 
Cryo-EM structure of CCR6 bound by PF-07054894 and OXM2
Method: single particle / : Wasilko DJ, Wu H

PDB-9pee: 
Cryo-EM structure of CCR6 bound by PF-07054894 and OXM2
Method: single particle / : Wasilko DJ, Wu H

EMDB-63903: 
FADD-DED filaments coordinate complex IIa assembly during TNF-induced apoptosis
Method: single particle / : Liu P, Luo D

EMDB-63933: 
FADD-DED filaments coordinate complex IIa assembly during TNF-induced apoptosis
Method: single particle / : Liu P, Luo D

PDB-9u6e: 
FADD-DED filaments coordinate complex IIa assembly during TNF-induced apoptosis
Method: single particle / : Liu P, Luo D

PDB-9u7a: 
FADD-DED filaments coordinate complex IIa assembly during TNF-induced apoptosis
Method: single particle / : Liu P, Luo D

EMDB-38039: 
Cryo-EM structure of CB2-G protein complex
Method: single particle / : Shen SY, Yang ZQ, Shao ZH

EMDB-36238: 
Cryo-EM structure of mClC-3 with AMP
Method: single particle / : Wan YZQ, Yang F

EMDB-36200: 
Cryo-EM structure of apo state mClC-3
Method: single particle / : Wan YZQ, Yang F

EMDB-36236: 
Cryo-EM structure of mClC-3 with ATP
Method: single particle / : Wan YZQ, Yang F

EMDB-36237: 
Cryo-EM structure of mClC-3 with ADP
Method: single particle / : Wan YZQ, Yang F

EMDB-36245: 
Cryo-EM structure of apo state mClC-3_I607T
Method: single particle / : Wan YZQ, Yang F

EMDB-36246: 
Cryo-EM structure of mClC-3_I607T with ATP
Method: single particle / : Wan YZQ, Yang F

EMDB-35767: 
Structure of the Mex67-Mtr2-3 heterodimer
Method: single particle / : Li ZQ, Chen SJ, Sui SF

EMDB-34638: 
Structure of the Mex67-Mtr2-1 heterodimer
Method: single particle / : Li ZQ, Chen SJ, Sui SF

EMDB-34640: 
Structure of Mex67-Mtr2-2 heterodimer
Method: single particle / : Li ZQ, Chen SJ, Sui SF

EMDB-34641: 
Structure of Crm1-RanGTP complex
Method: single particle / : Li ZQ, Chen SJ, Sui SF

EMDB-34725: 
NPC-trapped pre-60S particle
Method: single particle / : Li ZQ, Chen SJ, Sui SF

EMDB-35812: 
Bud20 interacts with CFNC
Method: single particle / : Li ZQ, Chen SJ

EMDB-33923: 
Cryo-EM structure of SARS-CoV-2 Omicron spike glycoprotein in complex with three neutralizing nanobody 3-2A2-4
Method: single particle / : Wang X, Zhang L, Ren Y, Li M

EMDB-15123: 
S. cerevisiae APC/C-Cdh1 complex
Method: single particle / : Barford D, Vazquez-Fernandez E, Zhang Z, Yang J

PDB-8a3t: 
S. cerevisiae APC/C-Cdh1 complex
Method: single particle / : Barford D, Vazquez-Fernandez E, Zhang Z, Yang J

EMDB-15201: 
S. cerevisiae apo phosphorylated APC/C
Method: single particle / : Barford D, Fernandez-Vazquez E, Zhang Z, Yang J

PDB-8a61: 
S. cerevisiae apo phosphorylated APC/C
Method: single particle / : Barford D, Fernandez-Vazquez E, Zhang Z, Yang J

EMDB-15199: 
S. cerevisiae apo unphosphorylated APC/C
Method: single particle / : Barford D, Fernandez-Vazquez E, Zhang Z, Yang J

PDB-8a5y: 
S. cerevisiae apo unphosphorylated APC/C.
Method: single particle / : Barford D, Fernandez-Vazquez E, Zhang Z, Yang J

EMDB-31502: 
S protein of SARS-CoV-2 in complex bound with P36-5D2(state2)
Method: single particle / : Zhang L, Wang X

EMDB-31503: 
S protein of SARS-CoV-2 in complex bound with P36-5D2 (state1)
Method: single particle / : Zhang L, Wang X
EMDB-20344: 
Polyclonal serum of rabbit C3 post 5th immunization in complex with heterologous Env trimer 45_01dG5 NFL TD 2CC+
Method: single particle / : Turner HL, Andrew AB

EMDB-20259: 
HIV Env BG505 NFL TD+ in complex with antibody E70 fragment antigen binding
Method: single particle / : Ozorowski G, Torres JL

EMDB-20260: 
HIV Env 16055 NFL TD 2CC+ in complex with antibody 1C2 fragment antigen binding
Method: single particle / : Ozorowski G, Torres JL
EMDB-20273: 
Negative-stain reconstruction of HIV-1 Env BG505 NFL CC+ trimer in complex with rabbit antibody E70 fragment antigen binding
Method: single particle / : Torres JL, Ozorowski G, Andrew AB
EMDB-20274: 
Negative stain reconstruction of HIV-1 Env 16055 NFL TD 2CC+ trimer in complex with rabbit antibody 1C2 fragment antigen binding
Method: single particle / : Torres JL, Ozorowski G, Andrew AB
EMDB-20279: 
Polyclonal Fabs from rabbit C3 post 5th immunization in complex with heterologous Env trimer 45_01dH5
Method: single particle / : Cottrell CA, Andrew AB

EMDB-20280: 
Polyclonal Fabs from rabbit C3 post 5th immunization in complex with heterologous Env trimer 45_01dH5
Method: single particle / : Cottrell CA, Andrew AB
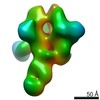
EMDB-20342: 
Polyclonal serum of rabbit C3 boost 5 in complex with heterologous Env trimer SC422 NFL TD CC+
Method: single particle / : Turner HL, Andrew AB
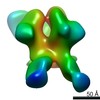
EMDB-20343: 
Polyclonal serum from rabbit C3 post 4th immunization in complex with heterologous Env trimer 45_01dG5 NFL TD 2CC+
Method: single particle / : Turner HL, Andrew AB
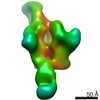
EMDB-20345: 
Polyclonal serum of rabbit A1 post 6th immunization in complex with homologous Env trimer 16055 NFL TD CC+
Method: single particle / : Turner HL, Andrew AB
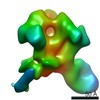
EMDB-20346: 
Polyclonal serum of rabbit A1 post 6th immunization in complex with heterologous Env trimer 45_01dG5 NFL TD 2CC+, class 1
Method: single particle / : Turner HL, Andrew AB
EMDB-20347: 
Polyclonal serum of rabbit A1 post 6th immunization in complex with heterologous Env trimer 45_01dG5 NFL TD 2CC+, class 2
Method: single particle / : Turner HL, Andrew AB

PDB-6p62: 
HIV Env BG505 NFL TD+ in complex with antibody E70 fragment antigen binding
Method: single particle / : Ozorowski G, Torres JL, Ward AB

PDB-6p65: 
HIV Env 16055 NFL TD 2CC+ in complex with antibody 1C2 fragment antigen binding
Method: single particle / : Ozorowski G, Torres JL, Ward AB

EMDB-9513: 
Cryo-EM map of the rabbit voltage-gated calcium channel Cav1.1 at 3.6 angstrom resolution
Method: single particle / : Wu JP, Yan Z
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
