+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
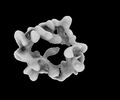
| Title | Structure of Crm1-RanGTP complex | |||||||||
 Map data Map data | Crm1-RanGTP complex half A map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nuclear export factor / NUCLEAR PROTEIN | |||||||||
| Biological species |  | |||||||||
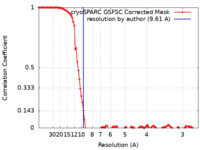
| Method | single particle reconstruction / cryo EM / Resolution: 9.61 Å | |||||||||
 Authors Authors | Li ZQ / Chen SJ / Sui SF | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Nuclear export of pre-60S particles through the nuclear pore complex. Authors: Zongqiang Li / Shuaijiabin Chen / Liang Zhao / Guoqiang Huang / Huiqin Xu / Xiaoyun Yang / Peiyi Wang / Ning Gao / Sen-Fang Sui /  Abstract: The nuclear pore complex (NPC) is the bidirectional gate that mediates the exchange of macromolecules or their assemblies between nucleus and cytoplasm. The assembly intermediates of the ribosomal ...The nuclear pore complex (NPC) is the bidirectional gate that mediates the exchange of macromolecules or their assemblies between nucleus and cytoplasm. The assembly intermediates of the ribosomal subunits, pre-60S and pre-40S particles, are among the largest cargoes of the NPC and the export of these gigantic ribonucleoproteins requires numerous export factors. Here we report the cryo-electron microscopy structure of native pre-60S particles trapped in the channel of yeast NPCs. In addition to known assembly factors, multiple factors with export functions are also included in the structure. These factors in general bind to either the flexible regions or subunit interface of the pre-60S particle, and virtually form many anchor sites for NPC binding. Through interactions with phenylalanine-glycine (FG) repeats from various nucleoporins of NPC, these factors collectively facilitate the passage of the pre-60S particle through the central FG repeat network of the NPC. Moreover, in silico analysis of the axial and radial distribution of pre-60S particles within the NPC shows that a single NPC can take up to four pre-60S particles simultaneously, and pre-60S particles are enriched in the inner ring regions close to the wall of the NPC with the solvent-exposed surface facing the centre of the nuclear pore. Our data suggest a translocation model for the export of pre-60S particles through the NPC. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34641.map.gz emd_34641.map.gz | 60.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34641-v30.xml emd-34641-v30.xml emd-34641.xml emd-34641.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34641_fsc.xml emd_34641_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_34641.png emd_34641.png | 33.9 KB | ||
| Others |  emd_34641_half_map_1.map.gz emd_34641_half_map_1.map.gz emd_34641_half_map_2.map.gz emd_34641_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34641 http://ftp.pdbj.org/pub/emdb/structures/EMD-34641 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34641 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34641 | HTTPS FTP |
-Validation report
| Summary document |  emd_34641_validation.pdf.gz emd_34641_validation.pdf.gz | 683.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34641_full_validation.pdf.gz emd_34641_full_validation.pdf.gz | 683 KB | Display | |
| Data in XML |  emd_34641_validation.xml.gz emd_34641_validation.xml.gz | 19.1 KB | Display | |
| Data in CIF |  emd_34641_validation.cif.gz emd_34641_validation.cif.gz | 24.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34641 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34641 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34641 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34641 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34641.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34641.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Crm1-RanGTP complex half A map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.668 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Crm1-RanGTP complex
| File | emd_34641_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Crm1-RanGTP complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Crm1-RanGTP complex half B map
| File | emd_34641_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Crm1-RanGTP complex half B map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Crm1-RanGTP complex
| Entire | Name: Crm1-RanGTP complex |
|---|---|
| Components |
|
-Supramolecule #1: Crm1-RanGTP complex
| Supramolecule | Name: Crm1-RanGTP complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Nuclear export factor
| Macromolecule | Name: Nuclear export factor / type: protein_or_peptide / ID: 1 Details: Author stated this map is a local refinement map from the peripheric region of the larger pre-60S particle (EMD-34725) with a high resolution. It is likely that the unmasked-calculated curve ...Details: Author stated this map is a local refinement map from the peripheric region of the larger pre-60S particle (EMD-34725) with a high resolution. It is likely that the unmasked-calculated curve has a higher resolution due to the influence of pre-60S particle. Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MEGILDFSND LDIALLDQVV STFYQGSGVQ QKQAQEILTK FQDNPDAWQK ADQILQFSTN PQSKFIALS ILDKLITRKW KLLPNDHRIG IRNFVVGMII SMCQDDEVFK TQKNLINKSD L TLVQILKQ EWPQNWPEFI PELIGSSSSS VNVCENNMIV LKLLSEEVFD ...String: MEGILDFSND LDIALLDQVV STFYQGSGVQ QKQAQEILTK FQDNPDAWQK ADQILQFSTN PQSKFIALS ILDKLITRKW KLLPNDHRIG IRNFVVGMII SMCQDDEVFK TQKNLINKSD L TLVQILKQ EWPQNWPEFI PELIGSSSSS VNVCENNMIV LKLLSEEVFD FSAEQMTQAK AL HLKNSMS KEFEQIFKLC FQVLEQGSSS SLIVATLESL LRYLHWIPYR YIYETNILEL LST KFMTSP DTRAITLKCL TEVSNLKIPQ DNDLIKRQTV LFFQNTLQQI ATSVMPVTAD LKAT YANAN GNDQSFLQDL AMFLTTYLAR NRALLESDES LRELLLNAHQ YLIQLSKIEE RELFK TTLD YWHNLVADLF YEVQRLPATE MSPLIQLSVG SQAISTGSGA LNPEYMKRFP LKKHIY EEI CSQLRLVIIE NMVRPEEVLV VENDEGEIVR EFVKESDTIQ LYKSEREVLV YLTHLNV ID TEEIMISKLA RQIDGSEWSW HNINTLSWAI GSISGTMSED TEKRFVVTVI KDLLDLTV K KRGKDNKAVV ASDIMYVVGQ YPRFLKAHWN FLRTVILKLF EFMHETHEGV QDMACDTFI KIVQKCKYHF VIQQPRESEP FIQTIIRDIQ KTTADLQPQQ VHTFYKACGI IISEERSVAE RNRLLSDLM QLPNMAWDTI VEQSTANPTL LLDSETVKII ANIIKTNVAV CTSMGADFYP Q LGHIYYNM LQLYRAVSSM ISAQVAAEGL IATKTPKVRG LRTIKKEILK LVETYISKAR NL DDVVKVL VEPLLNAVLE DYMNNVPDAR DAEVLNCMTT VVEKVGHMIP QGVILILQSV FEC TLDMIN KDFTEYPEHR VEFYKLLKVI NEKSFAAFLE LPPAAFKLFV DAICWAFKHN NRDV EVNGL QIALDLVKNI ERMGNVPFAN EFHKNYFFIF VSETFFVLTD SDHKSGFSKQ ALLLM KLIS LVYDNKISVP LYQEAEVPQG TSNQVYLSQY LANMLSNAFP HLTSEQIASF LSALTK QYK DLVVFKGTLR DFLVQIKEVG GDPTDYLFAE DKENALMEQN RLEREKAAKI GGLLKPS EL DD |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)