[English] 日本語
 Yorodumi
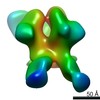
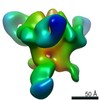
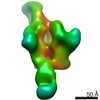
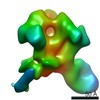
Yorodumi- PDB-6p62: HIV Env BG505 NFL TD+ in complex with antibody E70 fragment antig... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6p62 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | HIV Env BG505 NFL TD+ in complex with antibody E70 fragment antigen binding | |||||||||||||||
 Components Components |
| |||||||||||||||
 Keywords Keywords | VIRAL PROTEIN/immune system / HIV-1 / CD4 binding site / neutralizing antibody / rabbit antibody / VIRAL PROTEIN / VIRAL PROTEIN-immune system complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / membrane Similarity search - Function | |||||||||||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.57 Å | |||||||||||||||
 Authors Authors | Ozorowski, G. / Torres, J.L. / Ward, A.B. | |||||||||||||||
| Funding support |  United States, 4items United States, 4items
| |||||||||||||||
 Citation Citation |  Journal: Immunity / Year: 2019 Journal: Immunity / Year: 2019Title: Vaccination with Glycan-Modified HIV NFL Envelope Trimer-Liposomes Elicits Broadly Neutralizing Antibodies to Multiple Sites of Vulnerability. Authors: Viktoriya Dubrovskaya / Karen Tran / Gabriel Ozorowski / Javier Guenaga / Richard Wilson / Shridhar Bale / Christopher A Cottrell / Hannah L Turner / Gemma Seabright / Sijy O'Dell / Jonathan ...Authors: Viktoriya Dubrovskaya / Karen Tran / Gabriel Ozorowski / Javier Guenaga / Richard Wilson / Shridhar Bale / Christopher A Cottrell / Hannah L Turner / Gemma Seabright / Sijy O'Dell / Jonathan L Torres / Lifei Yang / Yu Feng / Daniel P Leaman / Néstor Vázquez Bernat / Tyler Liban / Mark Louder / Krisha McKee / Robert T Bailer / Arlette Movsesyan / Nicole A Doria-Rose / Marie Pancera / Gunilla B Karlsson Hedestam / Michael B Zwick / Max Crispin / John R Mascola / Andrew B Ward / Richard T Wyatt /    Abstract: The elicitation of broadly neutralizing antibodies (bNAbs) against the HIV-1 envelope glycoprotein (Env) trimer remains a major vaccine challenge. Most cross-conserved protein determinants are ...The elicitation of broadly neutralizing antibodies (bNAbs) against the HIV-1 envelope glycoprotein (Env) trimer remains a major vaccine challenge. Most cross-conserved protein determinants are occluded by self-N-glycan shielding, limiting B cell recognition of the underlying polypeptide surface. The exceptions to the contiguous glycan shield include the conserved receptor CD4 binding site (CD4bs) and glycoprotein (gp)41 elements proximal to the furin cleavage site. Accordingly, we performed heterologous trimer-liposome prime:boosting in rabbits to drive B cells specific for cross-conserved sites. To preferentially expose the CD4bs to B cells, we eliminated proximal N-glycans while maintaining the native-like state of the cleavage-independent NFL trimers, followed by gradual N-glycan restoration coupled with heterologous boosting. This approach successfully elicited CD4bs-directed, cross-neutralizing Abs, including one targeting a unique glycan-protein epitope and a bNAb (87% breadth) directed to the gp120:gp41 interface, both resolved by high-resolution cryoelectron microscopy. This study provides proof-of-principle immunogenicity toward eliciting bNAbs by vaccination. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6p62.cif.gz 6p62.cif.gz | 496.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6p62.ent.gz pdb6p62.ent.gz | 396.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6p62.json.gz 6p62.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/p6/6p62 https://data.pdbj.org/pub/pdb/validation_reports/p6/6p62 ftp://data.pdbj.org/pub/pdb/validation_reports/p6/6p62 ftp://data.pdbj.org/pub/pdb/validation_reports/p6/6p62 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  20259MC  6p65C  6pehC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 1 types, 3 molecules ABE
| #1: Protein | Mass: 74863.773 Da / Num. of mol.: 3 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Human immunodeficiency virus 1 / Cell line (production host): HEK293F / Production host: Human immunodeficiency virus 1 / Cell line (production host): HEK293F / Production host:  Homo sapiens (human) / References: UniProt: Q2N0S6*PLUS Homo sapiens (human) / References: UniProt: Q2N0S6*PLUS |
|---|
-Antibody , 2 types, 6 molecules HCFLDG
| #2: Antibody | Mass: 23527.469 Da / Num. of mol.: 3 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) Homo sapiens (human)#3: Antibody | Mass: 22414.732 Da / Num. of mol.: 3 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) Homo sapiens (human) |
|---|
-Sugars , 4 types, 45 molecules 
| #4: Polysaccharide | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source #5: Polysaccharide | Source method: isolated from a genetically manipulated source #6: Polysaccharide | Source method: isolated from a genetically manipulated source #7: Sugar | ChemComp-NAG / |
|---|
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: HIV-1 Env BG505 NFL TD+ in complex with rabbit monoclonal antibody E70 fragment antigen binding Type: COMPLEX / Entity ID: #1-#3 / Source: MULTIPLE SOURCES | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.57 MDa / Experimental value: NO | ||||||||||||||||
| Source (natural) |
| ||||||||||||||||
| Source (recombinant) | Organism:  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||
| Buffer solution | pH: 7.4 Details: LMNG added to sample shortly (< 5 minutes) before vitrification | ||||||||||||||||
| Buffer component |
| ||||||||||||||||
| Specimen | Conc.: 6 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||
| Specimen support | Grid material: COPPER / Grid mesh size: 400 divisions/in. / Grid type: C-flat-2/2 4C | ||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 283 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 29000 X / Nominal defocus max: 2000 nm / Nominal defocus min: 500 nm / Cs: 2.7 mm / C2 aperture diameter: 70 µm / Alignment procedure: COMA FREE |
| Specimen holder | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 12 sec. / Electron dose: 57 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Num. of real images: 870 |
| Image scans | Movie frames/image: 48 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 271757 | ||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C3 (3 fold cyclic) | ||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.57 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 49635 / Symmetry type: POINT |
 Movie
Movie Controller
Controller










 PDBj
PDBj






