-Search query
-Search result
Showing 1 - 50 of 4,789 items for (author: ton & w)

EMDB-42291: 
Structure of the human INTS9-INTS11-BRAT1 complex

EMDB-42292: 
Structure of the Drosophila IntS11-CG7044(dBRAT1) complex

EMDB-43814: 
Cryo-EM structure of the active Lactococcus lactis Csm bound to target in post-cleavage stage

EMDB-43815: 
Cryo-EM structure of the active Lactococcus lactis Csm bound to target in pre-cleavage stage

PDB-9ash: 
Cryo-EM structure of the active Lactococcus lactis Csm bound to target in post-cleavage stage

PDB-9asi: 
Cryo-EM structure of the active Lactococcus lactis Csm bound to target in pre-cleavage stage
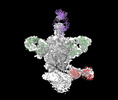
EMDB-44246: 
Cryo-EM structure of HIV-1 JRFL v6 Env in complex with vaccine-elicited, Membrane Proximal External Region (MPER) directed antibody DH1317.4.
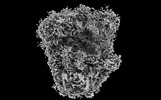
EMDB-41485: 
Subtomogram averaged consensus structure of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes
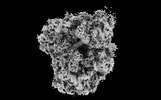
EMDB-41486: 
Subtomogram averaged decoding-1 state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes

EMDB-41487: 
Subtomogram averaged decoding-2 state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes

EMDB-41488: 
Subtomogram averaged classical iPRE state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes
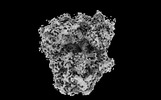
EMDB-41489: 
Subtomogram averaged rotated-1 PRE state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes

EMDB-41490: 
Subtomogram averaged rotated-2 PRE state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes

EMDB-41491: 
Subtomogram averaged translocation state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes
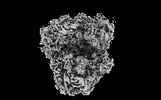
EMDB-41492: 
Subtomogram averaged POST state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes

EMDB-41493: 
Subtomogram averaged unloaded state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes
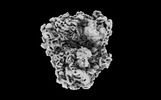
EMDB-41494: 
Subtomogram averaged non-rotated 80S ribosome of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes

PDB-8tpu: 
Subtomogram averaged consensus structure of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes

EMDB-39623: 
Cryo-EM structure of the retatrutide-bound human GCGR-Gs complex

PDB-8yw5: 
Cryo-EM structure of the retatrutide-bound human GCGR-Gs complex
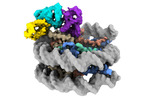
EMDB-38099: 
Cryo-EM structures of RNF168/UbcH5c-Ub in complex with H2AK13Ub nucleosomes determined by intein-based E2-Ub-NCP conjugation strategy
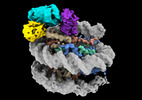
EMDB-38100: 
Cryo-EM structures of RNF168/UbcH5c-Ub/nucleosomes complex determined by activity-based chemical trapping strategy
EMDB-38101: 
Cryo-EM structures of RNF168/UbcH5c-Ub in complex with H2AK13Ub nucleosomes determined by activity-based chemical trapping strategy (adjacent H2AK13/15 dual-monoubiquitination)
EMDB-38102: 
Cryo-EM map of RNF168/UbcH5c-Ub/nucleosome determined by E2-Ub-NCP conjugation strategy
EMDB-36488: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Composite map)

EMDB-37212: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Receptor original map)
EMDB-37214: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Ligand/CCL7 focused map)

PDB-8jps: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Composite map)

EMDB-43746: 
Plasmodium falciparum 20S proteasome bound to an inhibitor

PDB-8w2f: 
Plasmodium falciparum 20S proteasome bound to an inhibitor

EMDB-44482: 
Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab
EMDB-44484: 
Cryo-EM structure of the HIV-1 BG505 IDL Env trimer in complex with 3BNC117 and 10-1074 Fabs
EMDB-44491: 
Cryo-EM structure of the HIV-1 WITO IDL Env trimer in complex with PGT122 Fab

PDB-9ber: 
Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab

PDB-9bew: 
Cryo-EM structure of the HIV-1 BG505 IDL Env trimer in complex with 3BNC117 and 10-1074 Fabs

PDB-9bf6: 
Cryo-EM structure of the HIV-1 WITO IDL Env trimer in complex with PGT122 Fab

EMDB-43139: 
SARS-CoV-2 Spike S2 bound to Fab 54043-5
EMDB-44232: 
GluA2 flip Q in complex with TARPgamma2 at pH8, consensus structure of LBD-TMD-TARPgamma2
EMDB-44233: 
GluA2 flip Q in complex with TARPgamma2 at pH8, consensus structure of TMD-TARPgamma2
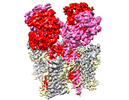
EMDB-44234: 
GluA2 flip Q in complex with TARPgamma2 at pH5, consensus structure of LBD-TMD-TARPgamma2

EMDB-44244: 
GluA2 flip Q in complex with TARPgamma2 at pH5, consensus structure of TMD-TARPgamma2
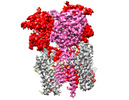
EMDB-44245: 
GluA2 flip Q in complex with TARPgamma2 at pH5, class23, structure of LBD-TMD-TARPgamma2
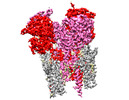
EMDB-44248: 
GluA2 flip Q in complex with TARPgamma2 at pH8, class1, structure of LBD-TMD-TARPgamma2
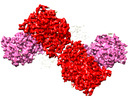
EMDB-44249: 
GluA2 flip Q in complex with TARPgamma2 at pH8, class1, structure of NTD
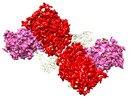
EMDB-44250: 
GluA2 flip Q in complex with TARPgamma2 at pH8, class12, structure of NTD
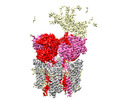
EMDB-44251: 
GluA2 flip Q in complex with TARPgamma2 at pH8, class12, structure of LBD-TMD-TARPgamma2

EMDB-18779: 
Structure of the non-mitochondrial citrate synthase from Ananas comosus

PDB-8qzp: 
Structure of the non-mitochondrial citrate synthase from Ananas comosus

EMDB-18973: 
Cryo-EM structure of Human SHMT1

PDB-8r7h: 
Cryo-EM structure of Human SHMT1
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
