-Search query
-Search result
Showing 1 - 50 of 439 items for (author: tani & y)

EMDB-36083: 
Cryo-EM structure of DDM1-nucleosome complex

EMDB-36084: 
Cryo-EM structure of nucleosome containing Arabidopsis thaliana histones

EMDB-36085: 
Cryo-EM structure of nucleosome containing Arabidopsis thaliana H2A.W

PDB-8j90: 
Cryo-EM structure of DDM1-nucleosome complex

PDB-8j91: 
Cryo-EM structure of nucleosome containing Arabidopsis thaliana histones

PDB-8j92: 
Cryo-EM structure of nucleosome containing Arabidopsis thaliana H2A.W

EMDB-60546: 
Flagellar central pair apparatus of Chlamydomonas reinhardtii wild type

EMDB-60565: 
Flagellar central pair apparatus of Chlamydomonas reinhardtii FAP47-deficient mutant.

EMDB-60566: 
Flagellar central pair apparatus of Chlamydomonas reinhardtii cpc1

EMDB-60567: 
Flagellar central pair apparatus of Chlamydomonas reinhardtii HYDIN-deficient mutant.

EMDB-60568: 
Flagellar central pair apparatus of Chlamydomonas reinhardtii with GFP-tag at the N-terminus of FAP47 protein.

EMDB-19926: 
CTE type I tau filament from vacuolar tauopathy

EMDB-19927: 
CTE type II tau filament from vacuolar tauopathy

EMDB-19928: 
CTE type II tau filament from vacuolar tauopathy

PDB-9erm: 
CTE type I tau filament from vacuolar tauopathy

PDB-9ern: 
CTE type II tau filament from vacuolar tauopathy

PDB-9ero: 
CTE type III tau filament from vacuolar tauopathy

EMDB-18664: 
Structure of the native microtubule lattice nucleated from the yeast spindle pole body

EMDB-18665: 
Structure of the native y-Tubulin Ring Complex (yTuRC) capping microtubule minus ends at the spindle pole body

EMDB-18666: 
Structure of the y-Tubulin Small Complex (yTuSC) as part of the native y-Tubulin Ring Complex (yTuRC) capping microtubule minus ends at the spindle pole body

PDB-8qv0: 
Structure of the native microtubule lattice nucleated from the yeast spindle pole body

PDB-8qv2: 
Structure of the native y-Tubulin Ring Complex (yTuRC) capping microtubule minus ends at the spindle pole body

PDB-8qv3: 
Structure of the y-Tubulin Small Complex (yTuSC) as part of the native y-Tubulin Ring Complex (yTuRC) capping microtubule minus ends at the spindle pole body

EMDB-39463: 
2.6A b-Galactosidase Structure Solved Using an Indirect Scintillator-Coupled CMOS Detector at 300 kV

EMDB-39481: 
2.08A Apoferritin Structure Solved Using an Indirect Scintillator-Coupled CMOS Detector at 300 kV

EMDB-17814: 
Structure of the human outer kinetochore KMN network complex

PDB-8ppr: 
Structure of the human outer kinetochore KMN network complex

EMDB-38291: 
Cryo-EM structure of human XKR8-basigin complex in lipid nanodisc

PDB-8xej: 
Cryo-EM structure of human XKR8-basigin complex in lipid nanodisc

EMDB-37465: 
Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by sucrose density

EMDB-37466: 
Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by Ca2+-DEAE

PDB-8wdu: 
Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by sucrose density

PDB-8wdv: 
Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by Ca2+-DEAE

EMDB-19194: 
In situ cryo-electron tomogram of an autophagosome in the projection of an iPSC-derived neuron #2

EMDB-19346: 
In situ cryo-electron tomogram of an autophagosome in the projection of an iPSC-derived neuron #1

EMDB-18387: 
FtsH1 protease from P.aeruginosa clone C in negative stain

EMDB-18388: 
FtsH2 protease from P.aeruginosa clone C in negative stain

EMDB-18389: 
P.aeruginosa clone C construct PaFtsH2-H1-link32 in negative stain

EMDB-36905: 
SARS-CoV-2 BA.1 RBD with UT28-RD

EMDB-36906: 
SARS-CoV-2 BA.1 spike with UT28-RD

PDB-8k5g: 
Structure of the SARS-CoV-2 BA.1 RBD with UT28-RD

PDB-8k5h: 
Structure of the SARS-CoV-2 BA.1 spike with UT28-RD

EMDB-16532: 
Structure of Tau filaments Type I from Subacute Sclerosing Panencephalitis

EMDB-16535: 
Structure of Tau filaments Type II from Subacute Sclerosing Panencephalitis
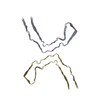
PDB-8caq: 
Structure of Tau filaments Type I from Subacute Sclerosing Panencephalitis

PDB-8cax: 
Structure of Tau filaments Type II from Subacute Sclerosing Panencephalitis

EMDB-26583: 
Cryo-EM structure of Antibody 12-16 in complex with prefusion SARS-CoV-2 Spike glycoprotein

PDB-7ukl: 
Cryo-EM structure of Antibody 12-16 in complex with prefusion SARS-CoV-2 Spike glycoprotein

EMDB-35912: 
Cryo-EM structure of the AsCas12f-sgRNA-target DNA ternary complex

EMDB-35926: 
Cryo-EM structure of the AsCas12f-YHAM-sgRNAS3-5v7-target DNA
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
