[English] 日本語
 Yorodumi
Yorodumi- EMDB-39463: 2.6A b-Galactosidase Structure Solved Using an Indirect Scintilla... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 2.6A b-Galactosidase Structure Solved Using an Indirect Scintillator-Coupled CMOS Detector at 300 kV | |||||||||
 Map data Map data | FSC-weighted, sharpend and masked map by PostProcess | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | b-galactosidase / CARBOHYDRATE | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.62 Å | |||||||||
 Authors Authors | Aramaki S / Yoshida Y / Tanihara T / Oyama K / Otsuki K / Terada Y / Matsunaga N / Ohdo S / Mayanagi K | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2026 Journal: J Struct Biol / Year: 2026Title: High-Resolution single particle analysis using a scintillator camera XF416 on CRYOARM300II at 300 kV. Authors: Shinji Aramaki / Tomohito Tanihara / Yuya Yoshida / Naoya Matsunaga / Shigehiro Ohdo / Kouta Mayanagi /  Abstract: The advent of direct electron detectors (DEDs) has driven a major breakthrough in cryo-electron microscopy (cryo-EM), particularly in single-particle analysis (SPA), establishing DEDs as essential ...The advent of direct electron detectors (DEDs) has driven a major breakthrough in cryo-electron microscopy (cryo-EM), particularly in single-particle analysis (SPA), establishing DEDs as essential tools for achieving near-atomic resolution. In this study, we re-evaluated the performance of the TVIPS TemCam-XF416, an indirect scintillator-coupled CMOS camera (scintillator camera). Using a JEOL CRYOARM 300II, we performed SPA on two well-established benchmark specimens, β-galactosidase and apoferritin, at a 300 kV acceleration voltage. The resulting reconstructions reached resolutions of 2.6 Å and 2.1 Å, respectively. Notably, the apoferritin map clearly resolves the central holes of aromatic side chains-a level of detail previously considered exclusive to DEDs. These results were achieved by implementing the latest standard reconstruction workflows, including motion correction and contrast transfer function refinement, underscoring the critical role of computational methods in attaining high-resolution structures. While scintillator cameras inherently exhibit a lower signal-to-noise ratio than DEDs, our findings with XF416 demonstrate that, with appropriate data collection and processing, such cameras can deliver near-atomic resolution structures. This work establishes a crucial technical benchmark for the scintillator camera evaluated in this study on a high-end 300 kV cryo-EM platform, demonstrating its capability to achieve resolutions suitable for many structural biology applications and providing an updated perspective on its performance capabilities. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39463.map.gz emd_39463.map.gz | 11.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39463-v30.xml emd-39463-v30.xml emd-39463.xml emd-39463.xml | 16.7 KB 16.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_39463_fsc.xml emd_39463_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_39463.png emd_39463.png | 113 KB | ||
| Masks |  emd_39463_msk_1.map emd_39463_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-39463.cif.gz emd-39463.cif.gz | 4.5 KB | ||
| Others |  emd_39463_half_map_1.map.gz emd_39463_half_map_1.map.gz emd_39463_half_map_2.map.gz emd_39463_half_map_2.map.gz | 57 MB 57 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39463 http://ftp.pdbj.org/pub/emdb/structures/EMD-39463 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39463 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39463 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_39463.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39463.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | FSC-weighted, sharpend and masked map by PostProcess | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.865 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_39463_msk_1.map emd_39463_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
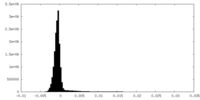
| Density Histograms |
-Half map: #1
| File | emd_39463_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
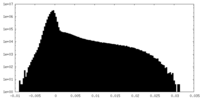
| Density Histograms |
-Half map: #2
| File | emd_39463_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
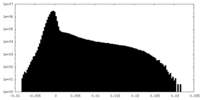
| Density Histograms |
- Sample components
Sample components
-Entire : beta-galactosidase
| Entire | Name: beta-galactosidase |
|---|---|
| Components |
|
-Supramolecule #1: beta-galactosidase
| Supramolecule | Name: beta-galactosidase / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 465 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.0 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Details: GloQube Plus | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: OTHER / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 9603 / Average exposure time: 2.3 sec. / Average electron dose: 107.0 e/Å2 / Details: TVIPS TemCam-XF416 (4k x 4k) |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.2 µm / Nominal magnification: 200000 |
| Sample stage | Specimen holder model: JEOL CRYOSPECPORTER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)