+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of DDM1-nucleosome complex | ||||||||||||||||||||||||||||||||||||||||||
 Map data Map data | Cryo-EM map of DDM1-nucleosome complex | ||||||||||||||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | Chromatin / Epigenetics / Histon variant / chromatin remodeler / NUCLEAR PROTEIN | ||||||||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationDNA-mediated transformation / retrotransposition / heterochromatin organization / chloroplast thylakoid / chromocenter / thylakoid / response to water deprivation / plasmodesma / plant-type vacuole / chloroplast stroma ...DNA-mediated transformation / retrotransposition / heterochromatin organization / chloroplast thylakoid / chromocenter / thylakoid / response to water deprivation / plasmodesma / plant-type vacuole / chloroplast stroma / plastid / DNA methylation-dependent constitutive heterochromatin formation / pericentric heterochromatin / heterochromatin / DNA helicase activity / epigenetic regulation of gene expression / chloroplast / structural constituent of chromatin / peroxisome / heterochromatin formation / nucleosome / DNA helicase / chromatin remodeling / protein heterodimerization activity / chromatin binding / nucleolus / ATP hydrolysis activity / DNA binding / extracellular region / ATP binding / nucleus / plasma membrane / cytosol Similarity search - Function | ||||||||||||||||||||||||||||||||||||||||||
| Biological species |  | ||||||||||||||||||||||||||||||||||||||||||
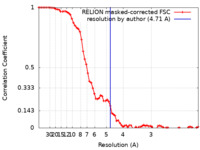
| Method | single particle reconstruction / cryo EM / Resolution: 4.71 Å | ||||||||||||||||||||||||||||||||||||||||||
 Authors Authors | Osakabe A / Takizawa Y / Horikoshi N / Hatazawa S / Berger F / Kurumizaka H / Kakutani T | ||||||||||||||||||||||||||||||||||||||||||
| Funding support |  Japan, Japan,  Austria, 13 items Austria, 13 items
| ||||||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Molecular and structural basis of the chromatin remodeling activity by Arabidopsis DDM1. Authors: Akihisa Osakabe / Yoshimasa Takizawa / Naoki Horikoshi / Suguru Hatazawa / Lumi Negishi / Shoko Sato / Frédéric Berger / Tetsuji Kakutani / Hitoshi Kurumizaka /   Abstract: The histone H2A variant H2A.W occupies transposons and thus prevents access to them in Arabidopsis thaliana. H2A.W is deposited by the chromatin remodeler DDM1, which also promotes the accessibility ...The histone H2A variant H2A.W occupies transposons and thus prevents access to them in Arabidopsis thaliana. H2A.W is deposited by the chromatin remodeler DDM1, which also promotes the accessibility of chromatin writers to heterochromatin by an unknown mechanism. To shed light on this question, we solve the cryo-EM structures of nucleosomes containing H2A and H2A.W, and the DDM1-H2A.W nucleosome complex. These structures show that the DNA end flexibility of the H2A nucleosome is higher than that of the H2A.W nucleosome. In the DDM1-H2A.W nucleosome complex, DDM1 binds to the N-terminal tail of H4 and the nucleosomal DNA and increases the DNA end flexibility of H2A.W nucleosomes. Based on these biochemical and structural results, we propose that DDM1 counters the low accessibility caused by nucleosomes containing H2A.W to enable the maintenance of repressive epigenetic marks on transposons and prevent their activity. | ||||||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36083.map.gz emd_36083.map.gz | 4.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36083-v30.xml emd-36083-v30.xml emd-36083.xml emd-36083.xml | 30.5 KB 30.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36083_fsc.xml emd_36083_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_36083.png emd_36083.png | 103.2 KB | ||
| Filedesc metadata |  emd-36083.cif.gz emd-36083.cif.gz | 7.9 KB | ||
| Others |  emd_36083_additional_1.map.gz emd_36083_additional_1.map.gz emd_36083_half_map_1.map.gz emd_36083_half_map_1.map.gz emd_36083_half_map_2.map.gz emd_36083_half_map_2.map.gz | 27.2 MB 23.4 MB 23.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36083 http://ftp.pdbj.org/pub/emdb/structures/EMD-36083 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36083 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36083 | HTTPS FTP |
-Related structure data
| Related structure data |  8j90MC  8j91C  8j92C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36083.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36083.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of DDM1-nucleosome complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Cryo-EM map of DDM1-nucleosome complex post-processed by DeepEMhancer
| File | emd_36083_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of DDM1-nucleosome complex post-processed by DeepEMhancer | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_36083_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_36083_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : DDM1-nucleosome complex
| Entire | Name: DDM1-nucleosome complex |
|---|---|
| Components |
|
-Supramolecule #1: DDM1-nucleosome complex
| Supramolecule | Name: DDM1-nucleosome complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 300 KDa |
-Macromolecule #1: Histone H3.1
| Macromolecule | Name: Histone H3.1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.583246 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSHMARTKQT ARKSTGGKAP RKQLATKAAR KSAPATGGVK KPHRFRPGTV ALREIRKYQK STELLIRKLP FQRLVREIAQ DFKTDLRFQ SSAVAALQEA AEAYLVGLFE DTNLCAIHAK RVTIMPKDIQ LARRIRGERA UniProtKB: Histone H3.1 |
-Macromolecule #2: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.718744 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSHMSGRGKG GKGLGKGGAK RHRKVLRDNI QGITKPAIRR LARRGGVKRI SGLIYEETRG VLKIFLENVI RDAVTYTEHA RRKTVTAMD VVYALKRQGR TLYGFGG UniProtKB: Histone H4 |
-Macromolecule #3: HTA6
| Macromolecule | Name: HTA6 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.280221 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSHMESTGKV KKAFGGRKPP GAPKTKSVSK SMKAGLQFPV GRITRFLKKG RYAQRLGGGA PVYMAAVLEY LAAEVLELAG NAARDNKKS RIIPRHLLLA IRNDEELGKL LSGVTIAHGG VLPNINSVLL PKKSATKPAE EKATKSPVKS PKKA UniProtKB: Probable histone H2A.7 |
-Macromolecule #4: HTB9
| Macromolecule | Name: HTB9 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.756738 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSHMAPRAEK KPAEKKPAAE KPVEEKSKAE KAPAEKKPKA GKKLPKEAGA GGDKKKKMKK KSVETYKIYI FKVLKQVHPD IGISSKAMG IMNSFINDIF EKLASESSKL ARYNKKPTIT SREIQTAVRL VLPGELAKHA VSEGTKAVTK FTSS UniProtKB: Histone H2B.6 |
-Macromolecule #7: ATP-dependent DNA helicase DDM1
| Macromolecule | Name: ATP-dependent DNA helicase DDM1 / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 87.050062 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPHMVSLRSR KVIPASEMVS DGKTEKDASG DSPTSVLNEE ENCEEKSVTV VEEEILLAKN GDSSLISEAM AQEEEQLLKL REDEEKANN AGSAVAPNLN ETQFTKLDEL LTQTQLYSEF LLEKMEDITI NGIESESQKA EPEKTGRGRK RKAASQYNNT K AKRAVAAM ...String: GPHMVSLRSR KVIPASEMVS DGKTEKDASG DSPTSVLNEE ENCEEKSVTV VEEEILLAKN GDSSLISEAM AQEEEQLLKL REDEEKANN AGSAVAPNLN ETQFTKLDEL LTQTQLYSEF LLEKMEDITI NGIESESQKA EPEKTGRGRK RKAASQYNNT K AKRAVAAM ISRSKEDGET INSDLTEEET VIKLQNELCP LLTGGQLKSY QLKGVKWLIS LWQNGLNGIL ADQMGLGKTI QT IGFLSHL KGNGLDGPYL VIAPLSTLSN WFNEIARFTP SINAIIYHGD KNQRDELRRK HMPKTVGPKF PIVITSYEVA MND AKRILR HYPWKYVVID EGHRLKNHKC KLLRELKHLK MDNKLLLTGT PLQNNLSELW SLLNFILPDI FTSHDEFESW FDFS EKNKN EATKEEEEKR RAQVVSKLHG ILRPFILRRM KCDVELSLPR KKEIIMYATM TDHQKKFQEH LVNNTLEAHL GENAI RGQG WKGKLNNLVI QLRKNCNHPD LLQGQIDGSY LYPPVEEIVG QCGKFRLLER LLVRLFANNH KVLIFSQWTK LLDIMD YYF SEKGFEVCRI DGSVKLDERR RQIKDFSDEK SSCSIFLLST RAGGLGINLT AADTCILYDS DWNPQMDLQA MDRCHRI GQ TKPVHVYRLS TAQSIETRVL KRAYSKLKLE HVVIGQGQFH QERAKSSTPL EEEDILALLK EDETAEDKLI QTDISDAD L DRLLDRSDLT ITAPGETQAA EAFPVKGPGW EVVLPSSGGM LSSLNS UniProtKB: ATP-dependent DNA helicase DDM1 |
-Macromolecule #5: DNA (169-MER)
| Macromolecule | Name: DNA (169-MER) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 51.922059 KDa |
| Sequence | String: (DA)(DT)(DC)(DG)(DG)(DA)(DC)(DC)(DC)(DT) (DA)(DT)(DC)(DG)(DC)(DG)(DA)(DG)(DC)(DC) (DA)(DG)(DG)(DC)(DC)(DT)(DG)(DA)(DG) (DA)(DA)(DT)(DC)(DC)(DG)(DG)(DT)(DG)(DC) (DC) (DG)(DA)(DG)(DG)(DC)(DC) ...String: (DA)(DT)(DC)(DG)(DG)(DA)(DC)(DC)(DC)(DT) (DA)(DT)(DC)(DG)(DC)(DG)(DA)(DG)(DC)(DC) (DA)(DG)(DG)(DC)(DC)(DT)(DG)(DA)(DG) (DA)(DA)(DT)(DC)(DC)(DG)(DG)(DT)(DG)(DC) (DC) (DG)(DA)(DG)(DG)(DC)(DC)(DG)(DC) (DT)(DC)(DA)(DA)(DT)(DT)(DG)(DG)(DT)(DC) (DG)(DT) (DA)(DG)(DA)(DC)(DA)(DG)(DC) (DT)(DC)(DT)(DA)(DG)(DC)(DA)(DC)(DC)(DG) (DC)(DT)(DT) (DA)(DA)(DA)(DC)(DG)(DC) (DA)(DC)(DG)(DT)(DA)(DC)(DG)(DC)(DG)(DC) (DT)(DG)(DT)(DC) (DC)(DC)(DC)(DC)(DG) (DC)(DG)(DT)(DT)(DT)(DT)(DA)(DA)(DC)(DC) (DG)(DC)(DC)(DA)(DA) (DG)(DG)(DG)(DG) (DA)(DT)(DT)(DA)(DC)(DT)(DC)(DC)(DC)(DT) (DA)(DG)(DT)(DC)(DT)(DC) (DC)(DA)(DG) (DG)(DC)(DA)(DC)(DG)(DT)(DG)(DT)(DC)(DA) (DG)(DA)(DT)(DA)(DT)(DA)(DT) (DA)(DC) (DA)(DT)(DC)(DC)(DG)(DA)(DT) |
-Macromolecule #6: DNA (169-MER)
| Macromolecule | Name: DNA (169-MER) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 52.424352 KDa |
| Sequence | String: (DA)(DT)(DC)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG)(DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DA) (DG)(DT)(DA)(DA)(DT)(DC) ...String: (DA)(DT)(DC)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG)(DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DA) (DG)(DT)(DA)(DA)(DT)(DC)(DC)(DC) (DC)(DT)(DT)(DG)(DG)(DC)(DG)(DG)(DT)(DT) (DA)(DA) (DA)(DA)(DC)(DG)(DC)(DG)(DG) (DG)(DG)(DG)(DA)(DC)(DA)(DG)(DC)(DG)(DC) (DG)(DT)(DA) (DC)(DG)(DT)(DG)(DC)(DG) (DT)(DT)(DT)(DA)(DA)(DG)(DC)(DG)(DG)(DT) (DG)(DC)(DT)(DA) (DG)(DA)(DG)(DC)(DT) (DG)(DT)(DC)(DT)(DA)(DC)(DG)(DA)(DC)(DC) (DA)(DA)(DT)(DT)(DG) (DA)(DG)(DC)(DG) (DG)(DC)(DC)(DT)(DC)(DG)(DG)(DC)(DA)(DC) (DC)(DG)(DG)(DA)(DT)(DT) (DC)(DT)(DC) (DA)(DG)(DG)(DC)(DC)(DT)(DG)(DG)(DC)(DT) (DC)(DG)(DC)(DG)(DA)(DT)(DA) (DG)(DG) (DG)(DT)(DC)(DC)(DG)(DA)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 25.0 µm / Nominal defocus min: 10.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)