-Search query
-Search result
Showing 1 - 50 of 100 items for (author: stella & s)

EMDB-43931: 
CryoEM structure of activated CRAF/MEK/14-3-3 complex with NST-628

EMDB-43932: 
Activated CRAF/MEK heterotetramer from focused refinement of CRAF/MEK/14-3-3 complex

PDB-9axa: 
CryoEM structure of activated CRAF/MEK/14-3-3 complex with NST-628

PDB-9axc: 
Activated CRAF/MEK heterotetramer from focused refinement of CRAF/MEK/14-3-3 complex

EMDB-15698: 
Cryo-EM structure of Cas12k-sgRNA binary complex (type V-K CRISPR-associated transposon)

PDB-8axb: 
Cryo-EM structure of Cas12k-sgRNA binary complex (type V-K CRISPR-associated transposon)

EMDB-15697: 
Cryo-EM structure of shCas12k-sgRNA-dsDNA ternary complex (type V-K CRISPR-associated transposon)

PDB-8axa: 
Cryo-EM structure of shCas12k-sgRNA-dsDNA ternary complex (type V-K CRISPR-associated transposon)
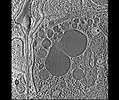
EMDB-28138: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
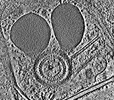
EMDB-28141: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
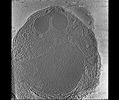
EMDB-28142: 
3D Reconstruction of Plasmodium falciparum (3D7) free merozoite
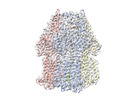
EMDB-17350: 
Single particle cryo-EM co-structure of Klebsiella pneumoniae AcrB with the BDM91288 efflux pump inhibitor at 2.97 Angstrom resolution

PDB-8p1i: 
Single particle cryo-EM co-structure of Klebsiella pneumoniae AcrB with the BDM91288 efflux pump inhibitor at 2.97 Angstrom resolution

EMDB-28125: 
Plasmodium falciparum merozoites apical 2-ring units

EMDB-28126: 
Toxoplasma apical rings

EMDB-28139: 
Toxoplasma gondii apical complex (ionophore stimulated)

EMDB-28140: 
Toxoplasma gondii apical complex (non-stimulated)

EMDB-15294: 
Cryo-EM structure of the strand transfer complex of the TnsB transposase (type V-K CRISPR-associated transposon)

EMDB-15344: 
Type V-K CAST TnsB bound to LTR-SR

PDB-8aa5: 
Cryo-EM structure of the strand transfer complex of the TnsB transposase (type V-K CRISPR-associated transposon)

EMDB-14622: 
Na+ - translocating ferredoxin: NAD+ reductase (Rnf) of C. tetanomorphum

PDB-7zc6: 
Na+ - translocating ferredoxin: NAD+ reductase (Rnf) of C. tetanomorphum

EMDB-26018: 
The symmetry-released subpellicular microtubule map from detergent-extracted Toxoplasma cells

EMDB-26019: 
Subpellicular microtubule from detergent-extract Toxoplasma gondii cells

EMDB-26020: 
The tubulin-based conoid from detergent-extract Toxoplasma gondii cells

PDB-7tnq: 
The symmetry-released subpellicular microtubule map from detergent-extracted Toxoplasma cells

PDB-7tns: 
Subpellicular microtubule from detergent-extract Toxoplasma gondii cells

PDB-7tnt: 
The tubulin-based conoid from detergent-extract Toxoplasma gondii cells

EMDB-23737: 
PolyQ40 oligomer tomogram at 2uM concentration

EMDB-23756: 
Tomogram of conditioned medium generated from PC12 14A2.6 cells showing mHTT assemblies
EMDB-23762: 
Tomogram of preformed liposomes incubated with polyQ40 oligomers at 2uM concentration

EMDB-23768: 
Tomogram of conditioned medium generated from PC12 14A2.6 cells showing mHTT assemblies (uncoated)

EMDB-26010: 
The symmetrized subpellicular microtubule map from intact Toxoplasma gondii cells

EMDB-26006: 
3D reconstruction of detergent-extract Toxoplasma gondii cells at the apical end

EMDB-26007: 
Three-dimensional organization of the apical complex in Toxoplasma tachyzoites

EMDB-26008: 
In situ average map of conoid with PCR from intact Toxoplasma gondii cells

EMDB-26009: 
The conoid segment from intact Toxoplasma gondii cells

EMDB-12827: 
Structure of the mini-RNA-guided endonuclease CRISPR-Cas_phi3

PDB-7odf: 
Structure of the mini-RNA-guided endonuclease CRISPR-Cas_phi3

EMDB-23122: 
Subtomogram average of the hexameric fungal plasma membrane proton pump Pma1 from protoplast membrane of Candida glabrata CBS138 strain

EMDB-23123: 
Subtomogram average of the hexameric fungal plasma membrane proton pump Pma1 from protoplast membrane of Candida glabrata KH238 strain

EMDB-22913: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the ACE2 protein decoy, CTC-445.2 (State 1)

EMDB-22914: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the ACE2 protein decoy, CTC-445.2 (State 2)

EMDB-22915: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the ACE2 protein decoy, CTC-445.2 (State 4)

EMDB-22916: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the ACE2 protein decoy, CTC-445.2 (State 4)

PDB-7kl9: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the ACE2 protein decoy, CTC-445.2 (State 4)

EMDB-10102: 
Type III-B Cmr-beta Cryo-EM structure of the Apo state

EMDB-10117: 
Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 1

EMDB-10119: 
Cryo-EM structure of the type III-B Cmr-beta complex bound to non-cognate target RNA

EMDB-10126: 
Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 2
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
