+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12827 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the mini-RNA-guided endonuclease CRISPR-Cas_phi3 | |||||||||||||||
 Map data Map data | map 3 different conformation | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | CRISPR-Cas / RNA-guided endonuclease / R-loop / RNA BINDING PROTEIN | |||||||||||||||
| Biological species |  Phage (virus) / synthetic construct (others) / Phage (virus) / synthetic construct (others) /  Phage #D (virus) Phage #D (virus) | |||||||||||||||
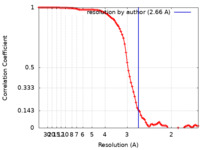
| Method | single particle reconstruction / cryo EM / Resolution: 2.66 Å | |||||||||||||||
 Authors Authors | Carabias del Rey A / Fugilsang A / Montoya G | |||||||||||||||
| Funding support |  Denmark, 4 items Denmark, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structure of the mini-RNA-guided endonuclease CRISPR-Cas12j3. Authors: Arturo Carabias / Anders Fuglsang / Piero Temperini / Tillmann Pape / Nicholas Sofos / Stefano Stella / Simon Erlendsson / Guillermo Montoya /   Abstract: CRISPR-Cas12j is a recently identified family of miniaturized RNA-guided endonucleases from phages. These ribonucleoproteins provide a compact scaffold gathering all key activities of a genome ...CRISPR-Cas12j is a recently identified family of miniaturized RNA-guided endonucleases from phages. These ribonucleoproteins provide a compact scaffold gathering all key activities of a genome editing tool. We provide the first structural insight into the Cas12j family by determining the cryoEM structure of Cas12j3/R-loop complex after DNA cleavage. The structure reveals the machinery for PAM recognition, hybrid assembly and DNA cleavage. The crRNA-DNA hybrid is directed to the stop domain that splits the hybrid, guiding the T-strand towards the catalytic site. The conserved RuvC insertion is anchored in the stop domain and interacts along the phosphate backbone of the crRNA in the hybrid. The assembly of a hybrid longer than 12-nt activates catalysis through key functional residues in the RuvC insertion. Our findings suggest why Cas12j unleashes unspecific ssDNA degradation after activation. A site-directed mutagenesis analysis supports the DNA cutting mechanism, providing new avenues to redesign CRISPR-Cas12j nucleases for genome editing. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12827.map.gz emd_12827.map.gz | 57.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12827-v30.xml emd-12827-v30.xml emd-12827.xml emd-12827.xml | 25.2 KB 25.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12827_fsc.xml emd_12827_fsc.xml | 11.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_12827.png emd_12827.png | 117.7 KB | ||
| Filedesc metadata |  emd-12827.cif.gz emd-12827.cif.gz | 7.4 KB | ||
| Others |  emd_12827_additional_1.map.gz emd_12827_additional_1.map.gz emd_12827_additional_2.map.gz emd_12827_additional_2.map.gz | 57.6 MB 54.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12827 http://ftp.pdbj.org/pub/emdb/structures/EMD-12827 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12827 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12827 | HTTPS FTP |
-Related structure data
| Related structure data |  7odfMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12827.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12827.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map 3 different conformation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: #1
| File | emd_12827_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
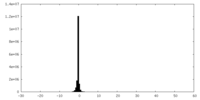
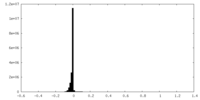
| Density Histograms |
-Additional map: map 2 different conformation
| File | emd_12827_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map 2 different conformation | ||||||||||||
| Projections & Slices |
| ||||||||||||
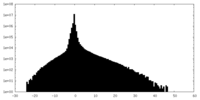
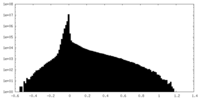
| Density Histograms |
- Sample components
Sample components
+Entire : cas_phi/R-loop complex
+Supramolecule #1: cas_phi/R-loop complex
+Supramolecule #2: Cas_phi3
+Supramolecule #3: DNA and RNA
+Macromolecule #1: Cas_phi3
+Macromolecule #2: DNA (5'-D(P*GP*TP*AP*TP*CP*CP*CP*AP*TP*TP*AP*CP*CP*AP*GP*CP*TP*GP...
+Macromolecule #3: DNA (5'-D(P*GP*TP*AP*AP*TP*TP*CP*AP*G)-3')
+Macromolecule #4: DNA (5'-D(P*GP*G)-3')
+Macromolecule #5: RNA (43-MER)
+Macromolecule #6: NICKEL (II) ION
+Macromolecule #7: ZINC ION
+Macromolecule #8: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Material: GOLD |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average exposure time: 1.05 sec. / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)