[English] 日本語
 Yorodumi
Yorodumi- EMDB-10126: Cryo-EM structure of the Type III-B Cmr-beta bound to cognate tar... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10126 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 2 | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | CRISPR-Cas / Effector complex / nuclease / cyclic oligo-adenylate synthase / ANTIVIRAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||
| Biological species |   Sulfolobus islandicus REY15A (archaea) / synthetic construct (others) / Sulfolobus islandicus REY15A (archaea) / synthetic construct (others) /   Sulfolobus islandicus (strain REY15A) (archaea) Sulfolobus islandicus (strain REY15A) (archaea) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.68 Å | ||||||||||||
 Authors Authors | Sofos N / Montoya G | ||||||||||||
| Funding support |  Denmark, 3 items Denmark, 3 items
| ||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2020 Journal: Mol Cell / Year: 2020Title: Structures of the Cmr-β Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas. Authors: Nicholas Sofos / Mingxia Feng / Stefano Stella / Tillmann Pape / Anders Fuglsang / Jinzhong Lin / Qihong Huang / Yingjun Li / Qunxin She / Guillermo Montoya /   Abstract: Cmr-β is a type III-B CRISPR-Cas complex that, upon target RNA recognition, unleashes a multifaceted immune response against invading genetic elements, including single-stranded DNA (ssDNA) ...Cmr-β is a type III-B CRISPR-Cas complex that, upon target RNA recognition, unleashes a multifaceted immune response against invading genetic elements, including single-stranded DNA (ssDNA) cleavage, cyclic oligoadenylate synthesis, and also a unique UA-specific single-stranded RNA (ssRNA) hydrolysis by the Cmr2 subunit. Here, we present the structure-function relationship of Cmr-β, unveiling how binding of the target RNA regulates the Cmr2 activities. Cryoelectron microscopy (cryo-EM) analysis revealed the unique subunit architecture of Cmr-β and captured the complex in different conformational stages of the immune response, including the non-cognate and cognate target-RNA-bound complexes. The binding of the target RNA induces a conformational change of Cmr2, which together with the complementation between the 5' tag in the CRISPR RNAs (crRNA) and the 3' antitag of the target RNA activate different configurations in a unique loop of the Cmr3 subunit, which acts as an allosteric sensor signaling the self- versus non-self-recognition. These findings highlight the diverse defense strategies of type III complexes. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10126.map.gz emd_10126.map.gz | 442.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10126-v30.xml emd-10126-v30.xml emd-10126.xml emd-10126.xml | 35.2 KB 35.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10126_fsc.xml emd_10126_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_10126.png emd_10126.png | 26.7 KB | ||
| Filedesc metadata |  emd-10126.cif.gz emd-10126.cif.gz | 8.9 KB | ||
| Others |  emd_10126_additional.map.gz emd_10126_additional.map.gz emd_10126_half_map_1.map.gz emd_10126_half_map_1.map.gz emd_10126_half_map_2.map.gz emd_10126_half_map_2.map.gz | 40.8 MB 109 MB 109 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10126 http://ftp.pdbj.org/pub/emdb/structures/EMD-10126 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10126 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10126 | HTTPS FTP |
-Related structure data
| Related structure data |  6s91MC  6s6bC  6s8bC  6s8eC  6sh8C  6shbC  6sicC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10126.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10126.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
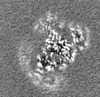
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
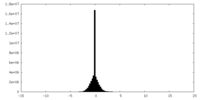
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Model-sharpened boxed map used for model building (Phenix)
| File | emd_10126_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Model-sharpened boxed map used for model building (Phenix) | ||||||||||||
| Projections & Slices |
| ||||||||||||
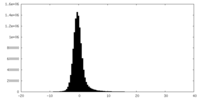
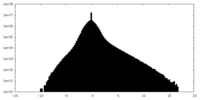
| Density Histograms |
-Half map: #1
| File | emd_10126_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
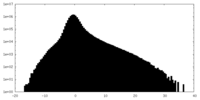
| Density Histograms |
-Half map: #2
| File | emd_10126_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
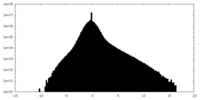
| Density Histograms |
- Sample components
Sample components
+Entire : Type III-B Cmr-beta ternary complex, cognate target RNA and AMPPn...
+Supramolecule #1: Type III-B Cmr-beta ternary complex, cognate target RNA and AMPPn...
+Supramolecule #2: CRISPR-associated proteins Cmr5, Cmr3, Cmr1, Cmr2, Cmr7
+Supramolecule #3: CRISPR-associated RAMP proteins Cmr4 and Cmr6, and crRNA
+Supramolecule #4: Cognate target RNA
+Macromolecule #1: CRISPR-associated protein, Cmr5 family
+Macromolecule #2: CRISPR-associated RAMP protein, Cmr4 family
+Macromolecule #3: CRISPR-associated protein, Cmr3 family
+Macromolecule #4: CRISPR-associated RAMP protein, Cmr6 family
+Macromolecule #5: Cmr1,CRISPR-associated RAMP protein, Cmr1 family
+Macromolecule #6: CRISPR-associated protein, Cmr2 family
+Macromolecule #9: CRISPR-associated protein Cmrx
+Macromolecule #7: Cognate target RNA
+Macromolecule #8: crRNA (48-MER)
+Macromolecule #10: ZINC ION
+Macromolecule #11: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
+Macromolecule #12: MANGANESE (II) ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6 Component:
| |||||||||||||||
| Grid | Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Details: 5 mA (Leica EM ACE200) | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 3-4 s blotting before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 3912 / Average exposure time: 40.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.1 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.7 µm / Nominal magnification: 96000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Target criteria: Cross-correlation coefficient |
|---|---|
| Output model |  PDB-6s91: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)