-Search query
-Search result
Showing 1 - 50 of 174 items for (author: smith & cl)

EMDB-44551: 
Map of eastern equine encephalitis virus q3 spike protein in complex with VLDLR without masked refinement

EMDB-42050: 
Structure of eastern equine encephalitis virus VLP in complex with VLDLR LA1

EMDB-42055: 
Structure of eastern equine encephalitis virus VLP unliganded quasi-threefold spike protein

EMDB-17295: 
Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '3 up' RBD conformation

PDB-8oyt: 
Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '3 up' RBD conformation

EMDB-19066: 
TREK2 in OGNG/CHS detergent micelle with biparatopic inhibitory nanobody Nb6158

EMDB-40825: 
10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10

PDB-8sx3: 
10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10

EMDB-17296: 
Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '2 up 1 down' RBD conformation

PDB-8oyu: 
Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '2 up 1 down' RBD conformation
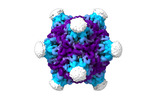
EMDB-19024: 
Structure of the PNMA2 capsid
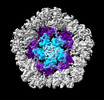
EMDB-19025: 
Structure of the five-fold capsomer of the PNMA2 capsid
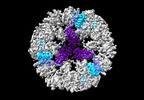
EMDB-19026: 
Structure of the three-fold capsomer of the PNMA2 capsid
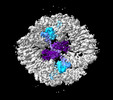
EMDB-19027: 
Structure of the two-fold capsomer of the PNMA2 capsid

PDB-8rb3: 
Structure of the PNMA2 capsid

PDB-8rb4: 
Structure of the five-fold capsomer of the PNMA2 capsid

PDB-8rb5: 
Structure of the three-fold capsomer of the PNMA2 capsid

PDB-8rb7: 
Structure of the two-fold capsomer of the PNMA2 capsid

EMDB-16110: 
Human Urea Transporter UT-A (N-Terminal Domain Model)

EMDB-16111: 
Map of Human Urea Transporter UT-A Collected with 0 and 30 Degree Tilts

EMDB-16112: 
Human Urea Transporter UT-B/UT1 in Complex with Inhibitor UTBinh-14

PDB-8blo: 
Human Urea Transporter UT-A (N-Terminal Domain Model)

PDB-8blp: 
Human Urea Transporter UT-B/UT1 in Complex with Inhibitor UTBinh-14

EMDB-41156: 
HCMV Trimer in complex with CS2it1p2_F7K Fab and CS4tt1p1_E3K Fab

EMDB-41157: 
Global reconstruction for HCMV Trimer in complex with CS2it1p2_F7K Fab and CS4tt1p1_E3K Fab

EMDB-41158: 
CS2it1p2_F7K local refinement for HCMV Trimer in complex with CS2it1p2_F7K Fab and CS4tt1p1_E3K Fab

EMDB-41160: 
CS4tt1p1_E3K local refinement for HCMV Trimer in complex with CS2it1p2_F7K Fab and CS4tt1p1_E3K Fab

EMDB-41161: 
gH base local refinement for HCMV Trimer in complex with CS2it1p2_F7K Fab and CS4tt1p1_E3K Fab

EMDB-41179: 
HCMV Pentamer in complex with CS2pt1p2_A10L Fab and CS3pt1p4_C1L Fab

EMDB-41180: 
Global reconstruction for HCMV Pentamer in complex with CS2pt1p2_A10L Fab and CS3pt1p4_C1L Fab

PDB-8tco: 
HCMV Trimer in complex with CS2it1p2_F7K Fab and CS4tt1p1_E3K Fab

PDB-8tea: 
HCMV Pentamer in complex with CS2pt1p2_A10L Fab and CS3pt1p4_C1L Fab

EMDB-41181: 
CS3pt1p4_C1L local refinement for HCMV Pentamer in complex with CS2pt1p2_A10L Fab and CS3pt1p4_C1L Fab

PDB-8sah: 
Huntingtin C-HEAT domain in complex with HAP40

EMDB-16364: 
The lipid linked oligosaccharide polymerase Wzy and its regulating co-polymerase Wzz form a complex in vivo and in vitro

PDB-8c0e: 
The lipid linked oligosaccharide polymerase Wzy and its regulating co-polymerase Wzz form a complex in vivo and in vitro
EMDB-27898: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with 2-oxohexadecyl-CoA
EMDB-27899: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with CoA and palmitoyl-LPA

PDB-8e4y: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with 2-oxohexadecyl-CoA

PDB-8e50: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with CoA and palmitoyl-LPA

EMDB-26475: 
Cryo-EM structure of PAPP-A in complex with IGFBP5

EMDB-27253: 
Cryo-EM structure of substrate unbound PAPP-A

PDB-7ufg: 
Cryo-EM structure of PAPP-A in complex with IGFBP5

PDB-8d8o: 
Cryo-EM structure of substrate unbound PAPP-A

EMDB-26738: 
Integrin alphaM/beta2 ectodomain in complex with adenylate cyclase toxin RTX751 and M1F5 Fab
EMDB-26739: 
Integrin alphaM/beta2 ectodomain

EMDB-27122: 
Global refinement for integrin alphaM/beta2 ectodomain in complex with adenylate cyclase toxin RTX751 and M1F5 Fab

EMDB-27123: 
Tailpiece local refinement for integrin alphaM/beta2 ectodomain in complex with adenylate cyclase toxin RTX751 and M1F5 Fab

EMDB-27124: 
Acylation domain local refinement for integrin alphaM/beta2 ectodomain in complex with adenylate cyclase toxin RTX751 and M1F5 Fab
EMDB-27125: 
Global refinement for integrin alphaM/beta2 ectodomain
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
