[English] 日本語
 Yorodumi
Yorodumi- EMDB-19066: TREK2 in OGNG/CHS detergent micelle with biparatopic inhibitory n... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | TREK2 in OGNG/CHS detergent micelle with biparatopic inhibitory nanobody Nb6158 | |||||||||
 Map data Map data | TREK2 with linked inhibitory nanobody in OGNG CHS detergent micelle at pH 7.5 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ion channel / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationTWIK related potassium channel (TREK) / cellular response to arachidonate / mechanosensitive potassium channel activity / Phase 4 - resting membrane potential / potassium ion leak channel activity / outward rectifier potassium channel activity / monoatomic ion channel complex / potassium channel activity / potassium ion transmembrane transport / signal transduction / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.37 Å | |||||||||
 Authors Authors | Smith KHM / Tucker SJ | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation. Authors: Karin E J Rödström / Alexander Cloake / Janina Sörmann / Agnese Baronina / Kathryn H M Smith / Ashley C W Pike / Jackie Ang / Peter Proks / Marcus Schewe / Ingelise Holland-Kaye / Simon R ...Authors: Karin E J Rödström / Alexander Cloake / Janina Sörmann / Agnese Baronina / Kathryn H M Smith / Ashley C W Pike / Jackie Ang / Peter Proks / Marcus Schewe / Ingelise Holland-Kaye / Simon R Bushell / Jenna Elliott / Els Pardon / Thomas Baukrowitz / Raymond J Owens / Simon Newstead / Jan Steyaert / Elisabeth P Carpenter / Stephen J Tucker /    Abstract: Potassium channels of the Two-Pore Domain (K2P) subfamily, KCNK1-KCNK18, play crucial roles in controlling the electrical activity of many different cell types and represent attractive therapeutic ...Potassium channels of the Two-Pore Domain (K2P) subfamily, KCNK1-KCNK18, play crucial roles in controlling the electrical activity of many different cell types and represent attractive therapeutic targets. However, the identification of highly selective small molecule drugs against these channels has been challenging due to the high degree of structural and functional conservation that exists not only between K2P channels, but across the whole K channel superfamily. To address the issue of selectivity, here we generate camelid antibody fragments (nanobodies) against the TREK-2 (KCNK10) K2P K channel and identify selective binders including several that directly modulate channel activity. X-ray crystallography and CryoEM data of these nanobodies in complex with TREK-2 also reveal insights into their mechanisms of activation and inhibition via binding to the extracellular loops and Cap domain, as well as their suitability for immunodetection. These structures facilitate design of a biparatropic inhibitory nanobody with markedly improved sensitivity. Together, these results provide important insights into TREK channel gating and provide an alternative, more selective approach to modulation of K2P channel activity via their extracellular domains. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19066.map.gz emd_19066.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19066-v30.xml emd-19066-v30.xml emd-19066.xml emd-19066.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19066_fsc.xml emd_19066_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_19066.png emd_19066.png | 33.1 KB | ||
| Masks |  emd_19066_msk_1.map emd_19066_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-19066.cif.gz emd-19066.cif.gz | 5.4 KB | ||
| Others |  emd_19066_half_map_1.map.gz emd_19066_half_map_1.map.gz emd_19066_half_map_2.map.gz emd_19066_half_map_2.map.gz | 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19066 http://ftp.pdbj.org/pub/emdb/structures/EMD-19066 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19066 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19066 | HTTPS FTP |
-Related structure data
| Related structure data |  8qz1C  8qz2C  8qz3C  8qz4C C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19066.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19066.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TREK2 with linked inhibitory nanobody in OGNG CHS detergent micelle at pH 7.5 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.94 Å | ||||||||||||||||||||||||||||||||||||
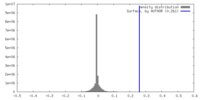
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19066_msk_1.map emd_19066_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
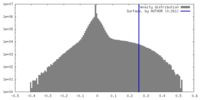
| Density Histograms |
-Half map: #2
| File | emd_19066_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
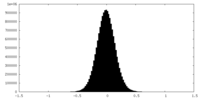
| Density Histograms |
-Half map: #1
| File | emd_19066_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
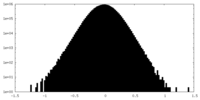
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of TREK2 and inhibitory nanobody Nb6158
| Entire | Name: Complex of TREK2 and inhibitory nanobody Nb6158 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of TREK2 and inhibitory nanobody Nb6158
| Supramolecule | Name: Complex of TREK2 and inhibitory nanobody Nb6158 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Macromolecule #1: TREK2
| Macromolecule | Name: TREK2 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MGLQTVMKWK TVVAIFVVVV VYLVTGGLVF RALEQPFESS QKNTIALEKA EFLRDHVCVS PQELETLIQH ALDADNAGVS PIGNSSNNSS HWDLGSAFFF AGTVITTIGY GNIAPSTEGG KIFCILYAIF GIPLFGFLLA GIGDQLGTIF GKSIARVEKV FRKKQVSQTK ...String: MGLQTVMKWK TVVAIFVVVV VYLVTGGLVF RALEQPFESS QKNTIALEKA EFLRDHVCVS PQELETLIQH ALDADNAGVS PIGNSSNNSS HWDLGSAFFF AGTVITTIGY GNIAPSTEGG KIFCILYAIF GIPLFGFLLA GIGDQLGTIF GKSIARVEKV FRKKQVSQTK IRVISTILFI LAGCIVFVTI PAVIFKYIEG WTALESIYFV VVTLTTVGFG DFVAGGNAGI NYREWYKPLV WFWILVGLAY FAAVLSMIGD WLRVLSKKTK EEVGEAENLY FQSHHHHHHH HHHDYKDDDD K UniProtKB: Potassium channel subfamily K member 10 |
-Macromolecule #2: Nb6158
| Macromolecule | Name: Nb6158 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLVESGGG LVQAGDSLRV SCAGSGFTFT SYGMGWFRQA PGKEREFVAS INWNSNTAYA DSVRGRFTIS RDNAESMMYL QMNSLKPEDT AVYYCAATRA YSKPRVDSRH YDYWGQGTQV TVSSGGGGSG GGGSGGGGSQ VQLVESGGGL VQAGGSLRLS CAASGRAGSG ...String: QVQLVESGGG LVQAGDSLRV SCAGSGFTFT SYGMGWFRQA PGKEREFVAS INWNSNTAYA DSVRGRFTIS RDNAESMMYL QMNSLKPEDT AVYYCAATRA YSKPRVDSRH YDYWGQGTQV TVSSGGGGSG GGGSGGGGSQ VQLVESGGGL VQAGGSLRLS CAASGRAGSG YAMGWFRQAP GKEREIVGAI SWSGDNTYYA DSVRGRVTIS RDYAQNTVYL QMNSLKPEDT AVYYCAADGR GNLRRGTAGR YVEYWGQGTQ VTVSSHHHHH HEPEA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.1 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)