-Search query
-Search result
Showing 1 - 50 of 214 items for (author: smith & ag)

EMDB-19066: 
TREK2 in OGNG/CHS detergent micelle with biparatopic inhibitory nanobody Nb6158
Method: single particle / : Smith KHM, Tucker SJ

EMDB-42977: 
Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 1)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

EMDB-43000: 
Cryo-EM structure of SNF2h-nucleosome complex (consensus structure)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

EMDB-43001: 
Cryo-EM structure of SNF2h-nucleosome complex (single-bound structure)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

EMDB-43002: 
Cryo-EM structure of doubly-bound SNF2h-nucleosome complex
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

EMDB-43003: 
Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 2)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

EMDB-43004: 
Cryo-EM structure of doubly-bound SNF2h-nucleosome complex (conformation 1)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

EMDB-43005: 
Cryo-EM structure of doubly-bound SNF2h-nucleosome complex (conformation 2)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

PDB-8v4y: 
Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 1)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

PDB-8v6v: 
Cryo-EM structure of doubly-bound SNF2h-nucleosome complex
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

PDB-8v7l: 
Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 2)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y
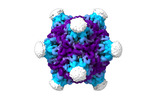
EMDB-19024: 
Structure of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
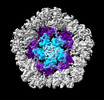
EMDB-19025: 
Structure of the five-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
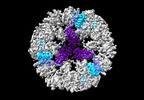
EMDB-19026: 
Structure of the three-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
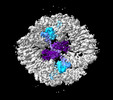
EMDB-19027: 
Structure of the two-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

EMDB-40334: 
Human OCT1 (Apo) in inward-open conformation
Method: single particle / : Zeng YC, Sobti M, Stewart AG

EMDB-40335: 
Human OCT1 bound to diltiazem in inward-open conformation
Method: single particle / : Zeng YC, Sobti M, Stewart AG

EMDB-40336: 
Human OCT1 bound to fenoterol in inward-open conformation
Method: single particle / : Zeng YC, Sobti M, Stewart AG

EMDB-40337: 
Human OCT1 bound to metformin in inward-open conformation
Method: single particle / : Zeng YC, Sobti M, Stewart AG

EMDB-40339: 
Human OCT1 bound to thiamine in inward-open conformation
Method: single particle / : Zeng YC, Sobti M, Stewart AG

EMDB-41153: 
Integrin alpha-v beta-8 in complex with minibinder B8_BP_dsulf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

EMDB-41154: 
Integrin alpha-v beta-6 in complex with minibinder B6_BP_dslf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

PDB-8tcf: 
Integrin alpha-v beta-8 in complex with minibinder B8_BP_dsulf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

PDB-8tcg: 
Integrin alpha-v beta-6 in complex with minibinder B6_BP_dslf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

EMDB-41158: 
CS2it1p2_F7K local refinement for HCMV Trimer in complex with CS2it1p2_F7K Fab and CS4tt1p1_E3K Fab
Method: single particle / : Goldsmith JA, McLellan JS

EMDB-41180: 
Global reconstruction for HCMV Pentamer in complex with CS2pt1p2_A10L Fab and CS3pt1p4_C1L Fab
Method: single particle / : Goldsmith JG, McLellan JS

EMDB-26809: 
KDM2A-nucleosome structure stabilized by H3K36C-UNC8015 covalent conjugate
Method: single particle / : Spangler CJ, Skrajna A, Foley CA, Budziszewski GR, Azzam DN, James LI, Frye SV, McGinty RK

EMDB-26810: 
KDM2B-nucleosome complex stabilized by H3K36C-UNC8015 covalent conjugate
Method: single particle / : Spangler CJ, Skrajna A, Foley CA, Budziszewski GR, Azzam DN, James LI, Frye SV, McGinty RK

PDB-7uv9: 
KDM2A-nucleosome structure stabilized by H3K36C-UNC8015 covalent conjugate
Method: single particle / : Spangler CJ, Skrajna A, Foley CA, Budziszewski GR, Azzam DN, James LI, Frye SV, McGinty RK

EMDB-29044: 
Structure of Zanidatamab bound to HER2
Method: single particle / : Worrall LJ, Atkinson CE, Sanches M, Dixit S, Strynadka NCJ

EMDB-28551: 
RMC-5552 in complex with mTORC1 and FKBP12
Method: single particle / : Tomlinson ACA, Yano JK

PDB-8era: 
RMC-5552 in complex with mTORC1 and FKBP12
Method: single particle / : Tomlinson ACA, Yano JK

EMDB-26862: 
CryoEM structure of the TIR domain from AbTir in complex with 3AD
Method: helical / : Li S, Nanson JD, Manik MK, Gu W, Landsberg MJ, Ve T, Kobe B

PDB-7uxu: 
CryoEM structure of the TIR domain from AbTir in complex with 3AD
Method: helical / : Li S, Nanson JD, Manik MK, Gu W, Landsberg MJ, Ve T, Kobe B

EMDB-26920: 
Staphylococcus epidermidis RP62a CRISPR effector subcomplex
Method: single particle / : Smith EM, Ferrell SH, Tokars VL, Mondragon A

EMDB-26921: 
Staphylococcus epidermidis RP62a CRISPR effector subcomplex with non-self target RNA bound
Method: single particle / : Smith EM, Ferrell SH, Tokars VL, Mondragon A

EMDB-26922: 
Staphylococcus epidermidis RP62A CRISPR effector complex with non-self target RNA 2
Method: single particle / : Smith EM, Ferrell SH, Tokars VL, Mondragon A

EMDB-26923: 
Staphylococcus epidermidis RP62a CRISPR tall effector complex
Method: single particle / : Smith EM, Ferrell SH, Tokars VL, Mondragon A

EMDB-26924: 
Staphylococcus epidermidis RP62a CRISPR tall effector complex with bound ATP
Method: single particle / : Smith EM, Ferrell SH, Tokars VL, Mondragon A

EMDB-26925: 
Staphylococcus epidermidis RP62a CRISPR short effector complex with self RNA target and ATP
Method: single particle / : Smith EM, Ferrell SH, Tokars VL, Mondragon A

EMDB-26926: 
Staphylococcus epidermidis RP62A CRISPR effector complex with non-self target RNA 1
Method: single particle / : Smith EM, Ferrell SH, Tokars VL, Mondragon A

EMDB-26927: 
Staphylococcus epidermidis RP62A CRISPR short effector complex
Method: single particle / : Smith EM, Ferrell SH, Tokars VL, Mondragon A

PDB-7uzw: 
Staphylococcus epidermidis RP62a CRISPR effector subcomplex
Method: single particle / : Smith EM, Ferrell SH, Tokars VL, Mondragon A

PDB-7uzx: 
Staphylococcus epidermidis RP62a CRISPR effector subcomplex with non-self target RNA bound
Method: single particle / : Smith EM, Ferrell SH, Tokars VL, Mondragon A

PDB-7uzy: 
Staphylococcus epidermidis RP62A CRISPR effector complex with non-self target RNA 2
Method: single particle / : Smith EM, Ferrell SH, Tokars VL, Mondragon A

PDB-7uzz: 
Staphylococcus epidermidis RP62a CRISPR tall effector complex
Method: single particle / : Smith EM, Ferrell SH, Tokars VL, Mondragon A

PDB-7v00: 
Staphylococcus epidermidis RP62a CRISPR tall effector complex with bound ATP
Method: single particle / : Smith EM, Ferrell SH, Tokars VL, Mondragon A

PDB-7v01: 
Staphylococcus epidermidis RP62a CRISPR short effector complex with self RNA target and ATP
Method: single particle / : Smith EM, Ferrell SH, Tokars VL, Mondragon A

PDB-7v02: 
Staphylococcus epidermidis RP62A CRISPR short effector complex
Method: single particle / : Smith EM, Ferrell SH, Tokars VL, Mondragon A

EMDB-26372: 
SARS-2 CoV 6P Mut7 in complex with Fab CC84.5
Method: single particle / : Torres JL, Ward AB
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
