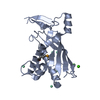
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Staphylococcus epidermidis RP62a CRISPR effector subcomplex | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Type IIIA CRISPR / effector complex / RNA BINDING PROTEIN / RNA BINDING PROTEIN-RNA complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationexonuclease activity / endonuclease activity / defense response to virus / hydrolase activity / RNA binding / ATP binding / metal ion binding Similarity search - Function | |||||||||||||||
| Biological species |  Staphylococcus epidermidis RP62A (bacteria) Staphylococcus epidermidis RP62A (bacteria) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.55 Å | |||||||||||||||
 Authors Authors | Smith EM / Ferrell SH / Tokars VL / Mondragon A | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2022 Journal: Structure / Year: 2022Title: Structures of an active type III-A CRISPR effector complex. Authors: Eric M Smith / Sé Ferrell / Valerie L Tokars / Alfonso Mondragón /  Abstract: Clustered regularly interspaced short palindromic repeats (CRISPR) and their CRISPR-associated proteins (Cas) provide many prokaryotes with an adaptive immune system against invading genetic material. ...Clustered regularly interspaced short palindromic repeats (CRISPR) and their CRISPR-associated proteins (Cas) provide many prokaryotes with an adaptive immune system against invading genetic material. Type III CRISPR systems are unique in that they can degrade both RNA and DNA. In response to invading nucleic acids, they produce cyclic oligoadenylates that act as secondary messengers, activating cellular nucleases that aid in the immune response. Here, we present seven single-particle cryo-EM structures of the type III-A Staphylococcus epidermidis CRISPR effector complex. The structures reveal the intact S. epidermidis effector complex in an apo, ATP-bound, cognate target RNA-bound, and non-cognate target RNA-bound states and illustrate how the effector complex binds and presents crRNA. The complexes bound to target RNA capture the type III-A effector complex in a post-RNA cleavage state. The ATP-bound structures give details about how ATP binds to Cas10 to facilitate cyclic oligoadenylate production. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26920.map.gz emd_26920.map.gz | 141.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26920-v30.xml emd-26920-v30.xml emd-26920.xml emd-26920.xml | 23.3 KB 23.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26920_fsc.xml emd_26920_fsc.xml | 14.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_26920.png emd_26920.png | 64.3 KB | ||
| Masks |  emd_26920_msk_1.map emd_26920_msk_1.map | 282.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26920.cif.gz emd-26920.cif.gz | 7 KB | ||
| Others |  emd_26920_half_map_1.map.gz emd_26920_half_map_1.map.gz emd_26920_half_map_2.map.gz emd_26920_half_map_2.map.gz | 262.5 MB 262.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26920 http://ftp.pdbj.org/pub/emdb/structures/EMD-26920 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26920 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26920 | HTTPS FTP |
-Related structure data
| Related structure data |  7uzwMC  7uzxC  7uzyC  7uzzC  7v00C  7v01C  7v02C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26920.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26920.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26920_msk_1.map emd_26920_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_26920_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_26920_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Staphylococcus epidermidis RP62a CRISPR effector subcomplex
| Entire | Name: Staphylococcus epidermidis RP62a CRISPR effector subcomplex |
|---|---|
| Components |
|
-Supramolecule #1: Staphylococcus epidermidis RP62a CRISPR effector subcomplex
| Supramolecule | Name: Staphylococcus epidermidis RP62a CRISPR effector subcomplex type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Staphylococcus epidermidis RP62A (bacteria) / Strain: RP62a Staphylococcus epidermidis RP62A (bacteria) / Strain: RP62a |
-Macromolecule #1: CRISPR system Cms endoribonuclease Csm3
| Macromolecule | Name: CRISPR system Cms endoribonuclease Csm3 / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus epidermidis RP62A (bacteria) / Strain: ATCC 35984 / RP62A Staphylococcus epidermidis RP62A (bacteria) / Strain: ATCC 35984 / RP62A |
| Molecular weight | Theoretical: 24.033975 KDa |
| Sequence | String: MYSKIKISGT IEVVTGLHIG GGGESSMIGA IDSPVVRDLQ TKLPIIPGSS IKGKMRNLLA KHFGLKMKQE SHNQDDERVL RLFGSSEKG NIQRARLQIS DAFFSEKTKE HFAQNDIAYT ETKFENTINR LTAVANPRQI ERVTRGSEFD FVFIYNVDEE S QVEDDFEN ...String: MYSKIKISGT IEVVTGLHIG GGGESSMIGA IDSPVVRDLQ TKLPIIPGSS IKGKMRNLLA KHFGLKMKQE SHNQDDERVL RLFGSSEKG NIQRARLQIS DAFFSEKTKE HFAQNDIAYT ETKFENTINR LTAVANPRQI ERVTRGSEFD FVFIYNVDEE S QVEDDFEN IEKAIHLLEN DYLGGGGTRG NGRIQFKDTN IETVVGEYDS TNLKIK UniProtKB: CRISPR system Cms endoribonuclease Csm3 |
-Macromolecule #2: CRISPR system Cms protein Csm4
| Macromolecule | Name: CRISPR system Cms protein Csm4 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus epidermidis RP62A (bacteria) / Strain: ATCC 35984 / RP62A Staphylococcus epidermidis RP62A (bacteria) / Strain: ATCC 35984 / RP62A |
| Molecular weight | Theoretical: 34.551938 KDa |
| Sequence | String: MTLATKVFKL SFKTPVHFGK KRLSDGEMTI TADTLFSALF IETLQLGKDT DWLLNDLIIS DTFPYENELY YLPKPLIKID SKEEDNHKA FKKLKYVPVH HYNQYLNGEL SAEDATDLND IFNIGYFSLQ TKVSLIAQET DSSADSEPYS VGTFTFEPEA G LYFIAKGS ...String: MTLATKVFKL SFKTPVHFGK KRLSDGEMTI TADTLFSALF IETLQLGKDT DWLLNDLIIS DTFPYENELY YLPKPLIKID SKEEDNHKA FKKLKYVPVH HYNQYLNGEL SAEDATDLND IFNIGYFSLQ TKVSLIAQET DSSADSEPYS VGTFTFEPEA G LYFIAKGS EETLDHLNNI MTALQYSGLG GKRNAGYGQF EYEIINNQQL SKLLNQNGKH SILLSTAMAK KEEIESALKE AR YILTKRS GFVQSTNYSE MLVKKSDFYS FSSGSVFKNI FNGDIFNVGH NGKHPVYRYA KPLWLEV UniProtKB: CRISPR system Cms protein Csm4 |
-Macromolecule #3: CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1...
| Macromolecule | Name: CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A) type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus epidermidis RP62A (bacteria) / Strain: ATCC 35984 / RP62A Staphylococcus epidermidis RP62A (bacteria) / Strain: ATCC 35984 / RP62A |
| Molecular weight | Theoretical: 87.685781 KDa |
| Sequence | String: MNKKNILMYG SLLHDIGKII YRSGDHTFSR GTHSKLGHQF LSQFSEFKDN EVLDNVAYHH YKELAKANLD NDNTAYITYI ADNIASGID RRDIIEEGDE EYEKQLFNFD KYTPLYSVFN IVNSEKLKQT NGKFKFSNES NIEYPKTENI QYSSGNYTTL M KDMSHDLE ...String: MNKKNILMYG SLLHDIGKII YRSGDHTFSR GTHSKLGHQF LSQFSEFKDN EVLDNVAYHH YKELAKANLD NDNTAYITYI ADNIASGID RRDIIEEGDE EYEKQLFNFD KYTPLYSVFN IVNSEKLKQT NGKFKFSNES NIEYPKTENI QYSSGNYTTL M KDMSHDLE HKLSIKEGTF PSLLQWTESL WQYVPSSTNK NQLIDISLYD HSRITCAIAS CIFDYLNENN IHNYKDELFS KY ENTKSFY QKEAFLLLSM DMSGIQDFIY NISGSKALKS LRSRSFYLEL MLEVIVDQLL ERLELARANL LYTGGGHAYL LVS NTDKVK KKITQFNNEL KKWFMSEFTT DLSLSMAFEK CSGDDLMNTS GNYRTIWRNV SSKLSDIKAH KYSAEDILKL NHFH SYGDR ECKECLRSDI DINDDGLCSI CEGIINISND LRDKSFFVLS ETGKLKMPFN KFISVIDYEE AEMLVQNNNQ VRIYS KNKP YIGIGISTNL WMCDYDYASQ NQDMREKGIG SYVDREEGVK RLGVVRADID NLGATFISGI PEKYNSISRT ATLSRQ LSL FFKYELNHLL ENYQITAIYS GGDDLFLIGA WDDIIEASIY INDKFKEFTL DKLTLSAGVG MFSGKYPVSK MAFETGR LE EAAKTGEKNQ ISLWLQEKVY NWDEFKKNIL EEKLLVLQQG FSQTDEHGKA FIYKMLALLR NNEAINIARL AYLLARSK M NEDFTSKIFN WAQNDKDKNQ LITALEYYIY QIREAD UniProtKB: CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A) |
-Macromolecule #4: crRNA
| Macromolecule | Name: crRNA / type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Staphylococcus epidermidis RP62A (bacteria) / Strain: ATCC 35984 / RP62A Staphylococcus epidermidis RP62A (bacteria) / Strain: ATCC 35984 / RP62A |
| Molecular weight | Theoretical: 13.862377 KDa |
| Sequence | String: ACGAGAACAC GUAUGCCGAA GUAUAUAAAU CAUCAGUACA AAG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. Details: cryo-EM grids were prepared by glow discharging for 10 seconds at 15 mA in a Pelco easiGlow glow discharger. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: 3 ul of the protein-RNA complex were added to the grid and allowed to incubate for 10 seconds. After incubation the grid was blotted in a Vitrobot Mark IV (FEI Thermo Fischer) (humidity 95% ...Details: 3 ul of the protein-RNA complex were added to the grid and allowed to incubate for 10 seconds. After incubation the grid was blotted in a Vitrobot Mark IV (FEI Thermo Fischer) (humidity 95% and temperature 4 C) for 3 seconds with a force of 0 before plunge freezing in liquid ethane.. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 2455 / Average electron dose: 49.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 44454 / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: OTHER | ||||||||||||
| Output model |  PDB-7uzw: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)