[English] 日本語
 Yorodumi
Yorodumi- PDB-7uzy: Staphylococcus epidermidis RP62A CRISPR effector complex with non... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7uzy | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
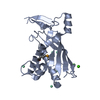
| Title | Staphylococcus epidermidis RP62A CRISPR effector complex with non-self target RNA 2 | |||||||||||||||
 Components Components |
| |||||||||||||||
 Keywords Keywords | HYDROLASE/RNA / Type IIIA CRISPR / effector complex / RNA binding protein / HYDROLASE-RNA complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationexonuclease activity / endonuclease activity / defense response to virus / hydrolase activity / RNA binding / ATP binding / metal ion binding Similarity search - Function | |||||||||||||||
| Biological species |  Staphylococcus epidermidis RP62A (bacteria) Staphylococcus epidermidis RP62A (bacteria) | |||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 4.05 Å | |||||||||||||||
 Authors Authors | Smith, E.M. / Ferrell, S.H. / Tokars, V.L. / Mondragon, A. | |||||||||||||||
| Funding support |  United States, 4items United States, 4items
| |||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2022 Journal: Structure / Year: 2022Title: Structures of an active type III-A CRISPR effector complex. Authors: Eric M Smith / Sé Ferrell / Valerie L Tokars / Alfonso Mondragón /  Abstract: Clustered regularly interspaced short palindromic repeats (CRISPR) and their CRISPR-associated proteins (Cas) provide many prokaryotes with an adaptive immune system against invading genetic material. ...Clustered regularly interspaced short palindromic repeats (CRISPR) and their CRISPR-associated proteins (Cas) provide many prokaryotes with an adaptive immune system against invading genetic material. Type III CRISPR systems are unique in that they can degrade both RNA and DNA. In response to invading nucleic acids, they produce cyclic oligoadenylates that act as secondary messengers, activating cellular nucleases that aid in the immune response. Here, we present seven single-particle cryo-EM structures of the type III-A Staphylococcus epidermidis CRISPR effector complex. The structures reveal the intact S. epidermidis effector complex in an apo, ATP-bound, cognate target RNA-bound, and non-cognate target RNA-bound states and illustrate how the effector complex binds and presents crRNA. The complexes bound to target RNA capture the type III-A effector complex in a post-RNA cleavage state. The ATP-bound structures give details about how ATP binds to Cas10 to facilitate cyclic oligoadenylate production. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7uzy.cif.gz 7uzy.cif.gz | 356.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7uzy.ent.gz pdb7uzy.ent.gz | 282.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7uzy.json.gz 7uzy.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/uz/7uzy https://data.pdbj.org/pub/pdb/validation_reports/uz/7uzy ftp://data.pdbj.org/pub/pdb/validation_reports/uz/7uzy ftp://data.pdbj.org/pub/pdb/validation_reports/uz/7uzy | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  26922MC  7uzwC  7uzxC  7uzzC  7v00C  7v01C  7v02C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-CRISPR system ... , 4 types, 9 molecules FJIKDCABH
| #1: Protein | Mass: 87685.781 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Staphylococcus epidermidis RP62A (bacteria) Staphylococcus epidermidis RP62A (bacteria)Strain: ATCC 35984 / RP62A / References: UniProt: Q5HK89 | ||||
|---|---|---|---|---|---|
| #2: Protein | Mass: 15436.794 Da / Num. of mol.: 3 / Source method: isolated from a natural source Source: (natural)  Staphylococcus epidermidis RP62A (bacteria) Staphylococcus epidermidis RP62A (bacteria)Strain: ATCC 35984 / RP62A / References: UniProt: Q5HK90 #3: Protein | Mass: 24033.975 Da / Num. of mol.: 4 / Source method: isolated from a natural source Source: (natural)  Staphylococcus epidermidis RP62A (bacteria) Staphylococcus epidermidis RP62A (bacteria)Strain: ATCC 35984 / RP62A / References: UniProt: Q5HK91 #4: Protein | | Mass: 34551.938 Da / Num. of mol.: 1 / Source method: isolated from a natural source Source: (natural)  Staphylococcus epidermidis RP62A (bacteria) Staphylococcus epidermidis RP62A (bacteria)Strain: ATCC 35984 / RP62A / References: UniProt: Q5HK92 |
-RNA chain , 2 types, 2 molecules GL
| #5: RNA chain | Mass: 11811.033 Da / Num. of mol.: 1 Fragment: Staphylococcus epidermidis RP62A CRISPR RNA: Repeat plus Spacer sequence 2 Source method: isolated from a natural source Source: (natural)  Staphylococcus epidermidis RP62A (bacteria) Staphylococcus epidermidis RP62A (bacteria)Strain: ATCC 35984 / RP62A |
|---|---|
| #6: RNA chain | Mass: 12789.641 Da / Num. of mol.: 1 / Fragment: CRISPR non-self RNA target / Source method: obtained synthetically Source: (synth.)  Staphylococcus epidermidis RP62A (bacteria) Staphylococcus epidermidis RP62A (bacteria) |
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Staphylococcus epidermidis RP62a CRISPR effector complex with non-self target RNA Type: COMPLEX / Entity ID: all / Source: MULTIPLE SOURCES | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Experimental value: NO | ||||||||||||||||||||
| Source (natural) | Organism:  | ||||||||||||||||||||
| Buffer solution | pH: 8 | ||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||
| Specimen | Conc.: 3.2 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||
| Specimen support | Details: cryo-EM grids were prepared by glow discharging for 10 seconds at 15 mA in a Pelco easiGlow glow discharger. Grid material: COPPER / Grid mesh size: 400 divisions/in. / Grid type: C-flat-1.2/1.3 | ||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 95 % / Chamber temperature: 277 K Details: 3 ul of the protein-RNA complex were added to the grid and allowed to incubate for 10 seconds. After incubation the grid was blotted in a Vitrobot Mark IV (FEI Thermo Fischer) (humidity 95% ...Details: 3 ul of the protein-RNA complex were added to the grid and allowed to incubate for 10 seconds. After incubation the grid was blotted in a Vitrobot Mark IV (FEI Thermo Fischer) (humidity 95% and temperature 4 C) for 3 seconds with a force of 0 before plunge freezing in liquid ethane. |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: TFS KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: OTHER FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: OTHER |
| Electron lens | Mode: BRIGHT FIELD / Calibrated magnification: 44454 X / Nominal defocus max: 2500 nm / Nominal defocus min: 1500 nm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 49.5 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) / Num. of real images: 2455 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 2368249 | ||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 4.05 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 42788 / Algorithm: FOURIER SPACE / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: OTHER / Space: REAL | ||||||||||||||||||||||||||||||||||||||||
| Atomic model building | 3D fitting-ID: 1 / Source name: PDB / Type: experimental model
| ||||||||||||||||||||||||||||||||||||||||
| Refinement | Cross valid method: THROUGHOUT | ||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 301.38 Å2 / Biso mean: 145.398 Å2 / Biso min: 30 Å2 |
 Movie
Movie Controller
Controller









 PDBj
PDBj