[English] 日本語
 Yorodumi
Yorodumi- EMDB-26862: CryoEM structure of the TIR domain from AbTir in complex with 3AD -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of the TIR domain from AbTir in complex with 3AD | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | 2' cADPR / NADase / Bacterial TIR / HYDROLASE | |||||||||
| Function / homology | NAD+ catabolic process / NAD+ nucleosidase activity / TIR domain / Toll - interleukin 1 - resistance / TIR domain profile. / Toll/interleukin-1 receptor homology (TIR) domain / Toll/interleukin-1 receptor homology (TIR) domain superfamily / signal transduction / Molecular chaperone Tir Function and homology information Function and homology information | |||||||||
| Biological species |  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) | |||||||||
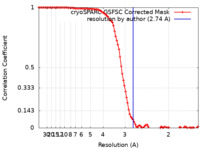
| Method | helical reconstruction / cryo EM / Resolution: 2.74 Å | |||||||||
 Authors Authors | Li S / Nanson JD / Manik MK / Gu W / Landsberg MJ / Ve T / Kobe B | |||||||||
| Funding support |  Australia, 2 items Australia, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling. Authors: Mohammad K Manik / Yun Shi / Sulin Li / Mark A Zaydman / Neha Damaraju / Samuel Eastman / Thomas G Smith / Weixi Gu / Veronika Masic / Tamim Mosaiab / James S Weagley / Steven J Hancock / ...Authors: Mohammad K Manik / Yun Shi / Sulin Li / Mark A Zaydman / Neha Damaraju / Samuel Eastman / Thomas G Smith / Weixi Gu / Veronika Masic / Tamim Mosaiab / James S Weagley / Steven J Hancock / Eduardo Vasquez / Lauren Hartley-Tassell / Nestoras Kargios / Natsumi Maruta / Bryan Y J Lim / Hayden Burdett / Michael J Landsberg / Mark A Schembri / Ivan Prokes / Lijiang Song / Murray Grant / Aaron DiAntonio / Jeffrey D Nanson / Ming Guo / Jeffrey Milbrandt / Thomas Ve / Bostjan Kobe /    Abstract: Cyclic adenosine diphosphate (ADP)-ribose (cADPR) isomers are signaling molecules produced by bacterial and plant Toll/interleukin-1 receptor (TIR) domains via nicotinamide adenine dinucleotide ...Cyclic adenosine diphosphate (ADP)-ribose (cADPR) isomers are signaling molecules produced by bacterial and plant Toll/interleukin-1 receptor (TIR) domains via nicotinamide adenine dinucleotide (oxidized form) (NAD) hydrolysis. We show that v-cADPR (2'cADPR) and v2-cADPR (3'cADPR) isomers are cyclized by O-glycosidic bond formation between the ribose moieties in ADPR. Structures of 2'cADPR-producing TIR domains reveal conformational changes that lead to an active assembly that resembles those of Toll-like receptor adaptor TIR domains. Mutagenesis reveals a conserved tryptophan that is essential for cyclization. We show that 3'cADPR is an activator of ThsA effector proteins from the bacterial antiphage defense system termed Thoeris and a suppressor of plant immunity when produced by the effector HopAM1. Collectively, our results reveal the molecular basis of cADPR isomer production and establish 3'cADPR in bacteria as an antiviral and plant immunity-suppressing signaling molecule. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26862.map.gz emd_26862.map.gz | 12.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26862-v30.xml emd-26862-v30.xml emd-26862.xml emd-26862.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26862_fsc.xml emd_26862_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_26862.png emd_26862.png | 191.4 KB | ||
| Filedesc metadata |  emd-26862.cif.gz emd-26862.cif.gz | 5.8 KB | ||
| Others |  emd_26862_half_map_1.map.gz emd_26862_half_map_1.map.gz emd_26862_half_map_2.map.gz emd_26862_half_map_2.map.gz | 95.6 MB 95.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26862 http://ftp.pdbj.org/pub/emdb/structures/EMD-26862 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26862 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26862 | HTTPS FTP |
-Related structure data
| Related structure data |  7uxuMC  7uwgC  7uxrC  7uxsC  7uxtC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26862.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26862.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A
| File | emd_26862_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
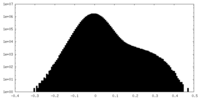
| Projections & Slices |
| ||||||||||||
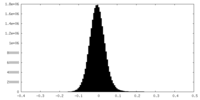
| Density Histograms |
-Half map: Half map B
| File | emd_26862_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
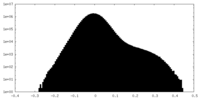
| Projections & Slices |
| ||||||||||||
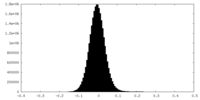
| Density Histograms |
- Sample components
Sample components
-Entire : Filament of the AbTIR TIR domain in complex with 3AD
| Entire | Name: Filament of the AbTIR TIR domain in complex with 3AD |
|---|---|
| Components |
|
-Supramolecule #1: Filament of the AbTIR TIR domain in complex with 3AD
| Supramolecule | Name: Filament of the AbTIR TIR domain in complex with 3AD / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) |
-Macromolecule #1: Molecular chaperone Tir
| Macromolecule | Name: Molecular chaperone Tir / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) |
| Molecular weight | Theoretical: 15.614538 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNAEYDLFIS HASEDKEDFV RPLAETLQQL GVNVWYDEFT LKVGDSLRQK IDSGLRNSKY GTVVLSTDFI KKDWTNYELD GLVAREMNG HKMILPIWHK ITKNDVLDYS PNLADKVALN TSVNSIEEIA HQLADVIL UniProtKB: Molecular chaperone Tir |
-Macromolecule #2: [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidany...
| Macromolecule | Name: [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(8-azanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl] ...Name: [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(8-azanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate type: ligand / ID: 2 / Number of copies: 4 / Formula: 1O4 |
|---|---|
| Molecular weight | Theoretical: 686.482 Da |
| Chemical component information | 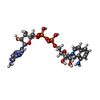 ChemComp-1O4: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil / Material: COPPER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 96 % / Chamber temperature: 281 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 0.1 µm |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)