+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
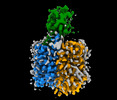
| Title | Human OCT1 (Apo) in inward-open conformation | |||||||||
 Map data Map data | Human OCT1 (Apo) in inward-open conformation | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | membrane protein / MFS / drug transporter / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationacetylcholine transport / O-acyl-L-carnitine transmembrane transport / pyrimidine nucleoside transmembrane transporter activity / serotonin transport / spermidine transmembrane transporter activity / epinephrine transport / purine-containing compound transmembrane transport / (R)-carnitine transmembrane transporter activity / putrescine transmembrane transporter activity / quaternary ammonium group transmembrane transporter activity ...acetylcholine transport / O-acyl-L-carnitine transmembrane transport / pyrimidine nucleoside transmembrane transporter activity / serotonin transport / spermidine transmembrane transporter activity / epinephrine transport / purine-containing compound transmembrane transport / (R)-carnitine transmembrane transporter activity / putrescine transmembrane transporter activity / quaternary ammonium group transmembrane transporter activity / acetylcholine transmembrane transporter activity / organic cation transport / SLC-mediated transport of organic cations / spermidine transport / quaternary ammonium group transport / : / dopamine uptake / putrescine transport / thiamine transmembrane transport / thiamine transmembrane transporter activity / thiamine transport / metanephric proximal tubule development / toxin transmembrane transporter activity / norepinephrine:sodium symporter activity / norepinephrine transport / dopamine:sodium symporter activity / prostaglandin transport / Norepinephrine Neurotransmitter Release Cycle / Abacavir transmembrane transport / prostaglandin transmembrane transporter activity / neurotransmitter transmembrane transporter activity / serotonin uptake / establishment or maintenance of transmembrane electrochemical gradient / dopamine transport / Neurotransmitter clearance / xenobiotic transport across blood-brain barrier / monoamine transmembrane transporter activity / monoamine transport / : / Ciprofloxacin ADME / SLC-mediated transport of neurotransmitters / cellular detoxification / xenobiotic transport / neurotransmitter transport / lateral plasma membrane / xenobiotic transmembrane transporter activity / transport across blood-brain barrier / xenobiotic metabolic process / basal plasma membrane / presynapse / basolateral plasma membrane / apical plasma membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
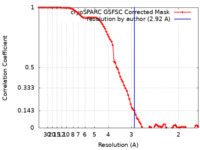
| Method | single particle reconstruction / cryo EM / Resolution: 2.92 Å | |||||||||
 Authors Authors | Zeng YC / Sobti M / Stewart AG | |||||||||
| Funding support |  Australia, 1 items Australia, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis of promiscuous substrate transport by Organic Cation Transporter 1. Authors: Yi C Zeng / Meghna Sobti / Ada Quinn / Nicola J Smith / Simon H J Brown / Jamie I Vandenberg / Renae M Ryan / Megan L O'Mara / Alastair G Stewart /  Abstract: Organic Cation Transporter 1 (OCT1) plays a crucial role in hepatic metabolism by mediating the uptake of a range of metabolites and drugs. Genetic variations can alter the efficacy and safety of ...Organic Cation Transporter 1 (OCT1) plays a crucial role in hepatic metabolism by mediating the uptake of a range of metabolites and drugs. Genetic variations can alter the efficacy and safety of compounds transported by OCT1, such as those used for cardiovascular, oncological, and psychological indications. Despite its importance in drug pharmacokinetics, the substrate selectivity and underlying structural mechanisms of OCT1 remain poorly understood. Here, we present cryo-EM structures of full-length human OCT1 in the inward-open conformation, both ligand-free and drug-bound, indicating the basis for its broad substrate recognition. Comparison of our structures with those of outward-open OCTs provides molecular insight into the alternating access mechanism of OCTs. We observe that hydrophobic gates stabilize the inward-facing conformation, whereas charge neutralization in the binding pocket facilitates the release of cationic substrates. These findings provide a framework for understanding the structural basis of the promiscuity of drug binding and substrate translocation in OCT1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40334.map.gz emd_40334.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40334-v30.xml emd-40334-v30.xml emd-40334.xml emd-40334.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40334_fsc.xml emd_40334_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_40334.png emd_40334.png | 86.3 KB | ||
| Filedesc metadata |  emd-40334.cif.gz emd-40334.cif.gz | 6.2 KB | ||
| Others |  emd_40334_half_map_1.map.gz emd_40334_half_map_1.map.gz emd_40334_half_map_2.map.gz emd_40334_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40334 http://ftp.pdbj.org/pub/emdb/structures/EMD-40334 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40334 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40334 | HTTPS FTP |
-Validation report
| Summary document |  emd_40334_validation.pdf.gz emd_40334_validation.pdf.gz | 760.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40334_full_validation.pdf.gz emd_40334_full_validation.pdf.gz | 759.7 KB | Display | |
| Data in XML |  emd_40334_validation.xml.gz emd_40334_validation.xml.gz | 16.2 KB | Display | |
| Data in CIF |  emd_40334_validation.cif.gz emd_40334_validation.cif.gz | 21 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40334 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40334 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40334 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40334 | HTTPS FTP |
-Related structure data
| Related structure data |  8sc1MC  8sc2C  8sc3C  8sc4C  8sc6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40334.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40334.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human OCT1 (Apo) in inward-open conformation | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
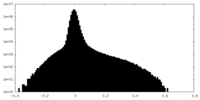
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_40334_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
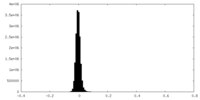
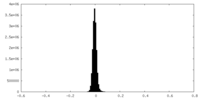
| Density Histograms |
-Half map: Half Map 2
| File | emd_40334_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
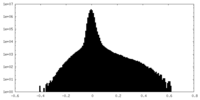
| Density Histograms |
- Sample components
Sample components
-Entire : Human OCT1
| Entire | Name: Human OCT1 |
|---|---|
| Components |
|
-Supramolecule #1: Human OCT1
| Supramolecule | Name: Human OCT1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 22 member 1
| Macromolecule | Name: Solute carrier family 22 member 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 61.200664 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPTVDDILEQ VGESGWFQKQ AFLILCLLSA AFAPICVGIV FLGFTPDHHC QSPGVAELSQ RCGWSPAEEL NYTVPGLGPA GEAFLGQCR RYEVDWNQSA LSCVDPLASL ATNRSHLPLG PCQDGWVYDT PGSSIVTEFN LVCADSWKLD LFQSCLNAGF L FGSLGVGY ...String: MPTVDDILEQ VGESGWFQKQ AFLILCLLSA AFAPICVGIV FLGFTPDHHC QSPGVAELSQ RCGWSPAEEL NYTVPGLGPA GEAFLGQCR RYEVDWNQSA LSCVDPLASL ATNRSHLPLG PCQDGWVYDT PGSSIVTEFN LVCADSWKLD LFQSCLNAGF L FGSLGVGY FADRFGRKLC LLGTVLVNAV SGVLMAFSPN YMSMLLFRLL QGLVSKGNWM AGYTLITEFV GSGSRRTVAI MY QMAFTVG LVALTGLAYA LPHWRWLQLA VSLPTFLFLL YYWCVPESPR WLLSQKRNTE AIKIMDHIAQ KNGKLPPADL KML SLEEDV TEKLSPSFAD LFRTPRLRKR TFILMYLWFT DSVLYQGLIL HMGATSGNLY LDFLYSALVE IPGAFIALIT IDRV GRIYP MAMSNLLAGA ACLVMIFISP DLHWLNIIIM CVGRMGITIA IQMICLVNAE LYPTFVRNLG VMVCSSLCDI GGIIT PFIV FRLREVWQAL PLILFAVLGL LAAGVTLLLP ETKGVALPET MKDAENLGRK AKPKENTIYL KVQTSEPSGT UniProtKB: Solute carrier family 22 member 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 88.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)