-Search query
-Search result
Showing 1 - 50 of 192 items for (author: olia & as)

EMDB-44482: 
Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab
EMDB-44484: 
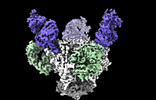
Cryo-EM structure of the HIV-1 BG505 IDL Env trimer in complex with 3BNC117 and 10-1074 Fabs
EMDB-44491: 
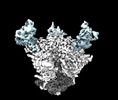
Cryo-EM structure of the HIV-1 WITO IDL Env trimer in complex with PGT122 Fab

PDB-9ber: 
Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab

PDB-9bew: 
Cryo-EM structure of the HIV-1 BG505 IDL Env trimer in complex with 3BNC117 and 10-1074 Fabs

PDB-9bf6: 
Cryo-EM structure of the HIV-1 WITO IDL Env trimer in complex with PGT122 Fab

EMDB-41569: 
Cryo-EM structure of HmAb64 scFv in complex with CNE40 SOSIP trimer

EMDB-43529: 
L5A7 Fab bound to Indonesia2005 Hemagglutinin

EMDB-43545: 
L5A7 Fab bound to 28H6E11 anti-idiotype Fab

PDB-8vue: 
L5A7 Fab bound to Indonesia2005 Hemagglutinin

PDB-8vuz: 
L5A7 Fab bound to 28H6E11 anti-idiotype Fab

EMDB-18609: 
Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 8.0

EMDB-18610: 
Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 4.3

PDB-8qqz: 
Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 8.0

PDB-8qr0: 
Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 4.3

EMDB-18381: 
UFL1 E3 ligase bound 60S ribosome

EMDB-18382: 
UFL1 E3 ligase bound 60S ribosome

PDB-8qfc: 
UFL1 E3 ligase bound 60S ribosome

PDB-8qfd: 
UFL1 E3 ligase bound 60S ribosome
EMDB-28617: 
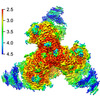
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01 FAB

EMDB-28618: 
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-COMBO1 FAB

EMDB-28619: 
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-MM28 FAB

PDB-8euu: 
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01 FAB

PDB-8euv: 
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-COMBO1 FAB

PDB-8euw: 
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-MM28 FAB

EMDB-28910: 
Glycan-Base ConC Env Trimer

PDB-8f7t: 
Glycan-Base ConC Env Trimer

EMDB-29725: 
Vaccine-elicited human antibody 2C06 in complex with HIV-1 envelope trimer BG505 DS-SOSIP

EMDB-29731: 
Vaccine-elicited human antibody 2C09 in complex with HIV-1 envelope trimer BG505 DS-SOSIP

PDB-8g4m: 
Vaccine-elicited human antibody 2C06 in complex with HIV-1 envelope trimer BG505 DS-SOSIP

PDB-8g4t: 
Vaccine-elicited human antibody 2C09 in complex with HIV-1 envelope trimer BG505 DS-SOSIP

EMDB-29396: 
Antibody vFP53.02 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
EMDB-29836: 
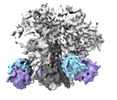
vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer

EMDB-29880: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1)

EMDB-29881: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 2)

EMDB-29882: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 3)

EMDB-29905: 
vFP48.02 Fab in complex with BG505 DS-SOSIP Env trimer

PDB-8fr6: 
Antibody vFP53.02 in complex with HIV-1 envelope trimer BG505 DS-SOSIP

PDB-8g85: 
vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer

PDB-8g9w: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1)

PDB-8g9x: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 2)

PDB-8g9y: 
Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 3)

PDB-8gas: 
vFP48.02 Fab in complex with BG505 DS-SOSIP Env trimer

EMDB-26157: 
Cryo-EM structure of BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9

PDB-7txd: 
Cryo-EM structure of BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9
EMDB-29209: 
Structure of Bispecific CAP256V2LS-J3 Fab in complex with BG505 DS-SOSIP.664

PDB-8fis: 
Structure of Bispecific CAP256V2LS-J3 Fab in complex with BG505 DS-SOSIP.664

EMDB-26200: 
Cryo-EM structure of SARS-CoV-2 spike in complex with FSR22, an anti-SARS-CoV-2 DARPin

EMDB-26201: 
Cryo-EM structure of SARS-CoV-2 spike in complex with FSR22, an anti-SARS-CoV-2 DARPin (Local refinement of FSR22 and RBD)
EMDB-27749: 
Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR22, and two antibody Fabs, S309 and CR3022
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
