[English] 日本語
 Yorodumi
Yorodumi- EMDB-41569: Cryo-EM structure of HmAb64 scFv in complex with CNE40 SOSIP trimer -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of HmAb64 scFv in complex with CNE40 SOSIP trimer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV / Env / glycoprotein / mAb / IMMUNE SYSTEM / IMMUNE SYSTEM-VIRAL PROTEIN complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.74 Å | |||||||||
 Authors Authors | Chan K-W / Kong XP | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Human CD4-binding site antibody elicited by polyvalent DNA prime-protein boost vaccine neutralizes cross-clade tier-2-HIV strains. Authors: Shixia Wang / Kun-Wei Chan / Danlan Wei / Xiuwen Ma / Shuying Liu / Guangnan Hu / Saeyoung Park / Ruimin Pan / Ying Gu / Alexandra F Nazzari / Adam S Olia / Kai Xu / Bob C Lin / Mark K ...Authors: Shixia Wang / Kun-Wei Chan / Danlan Wei / Xiuwen Ma / Shuying Liu / Guangnan Hu / Saeyoung Park / Ruimin Pan / Ying Gu / Alexandra F Nazzari / Adam S Olia / Kai Xu / Bob C Lin / Mark K Louder / Krisha McKee / Nicole A Doria-Rose / David Montefiori / Michael S Seaman / Tongqing Zhou / Peter D Kwong / James Arthos / Xiang-Peng Kong / Shan Lu /  Abstract: The vaccine elicitation of HIV tier-2-neutralization antibodies has been a challenge. Here, we report the isolation and characterization of a CD4-binding site (CD4bs) specific monoclonal antibody, ...The vaccine elicitation of HIV tier-2-neutralization antibodies has been a challenge. Here, we report the isolation and characterization of a CD4-binding site (CD4bs) specific monoclonal antibody, HmAb64, from a human volunteer immunized with a polyvalent DNA prime-protein boost HIV vaccine. HmAb64 is derived from heavy chain variable germline gene IGHV1-18 and light chain germline gene IGKV1-39. It has a third heavy chain complementarity-determining region (CDR H3) of 15 amino acids. On a cross-clade panel of 208 HIV-1 pseudo-virus strains, HmAb64 neutralized 20 (10%), including tier-2 strains from clades B, BC, C, and G. The cryo-EM structure of the antigen-binding fragment of HmAb64 in complex with a CNE40 SOSIP trimer revealed details of its recognition; HmAb64 uses both heavy and light CDR3s to recognize the CD4-binding loop, a critical component of the CD4bs. This study demonstrates that a gp120-based vaccine can elicit antibodies capable of tier 2-HIV neutralization. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41569.map.gz emd_41569.map.gz | 229.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41569-v30.xml emd-41569-v30.xml emd-41569.xml emd-41569.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41569.png emd_41569.png | 75.6 KB | ||
| Filedesc metadata |  emd-41569.cif.gz emd-41569.cif.gz | 6.7 KB | ||
| Others |  emd_41569_half_map_1.map.gz emd_41569_half_map_1.map.gz emd_41569_half_map_2.map.gz emd_41569_half_map_2.map.gz | 226.1 MB 226.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41569 http://ftp.pdbj.org/pub/emdb/structures/EMD-41569 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41569 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41569 | HTTPS FTP |
-Related structure data
| Related structure data |  8tr3MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41569.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41569.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.852 Å | ||||||||||||||||||||||||||||||||||||
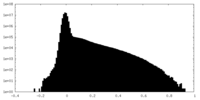
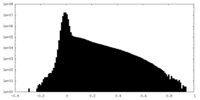
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_41569_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
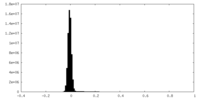
| Density Histograms |
-Half map: #1
| File | emd_41569_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
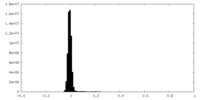
| Density Histograms |
- Sample components
Sample components
-Entire : 3D structure of HmAb64 Fv in complex with CNE40 SOSIP trimer
| Entire | Name: 3D structure of HmAb64 Fv in complex with CNE40 SOSIP trimer |
|---|---|
| Components |
|
-Supramolecule #1: 3D structure of HmAb64 Fv in complex with CNE40 SOSIP trimer
| Supramolecule | Name: 3D structure of HmAb64 Fv in complex with CNE40 SOSIP trimer type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|
-Supramolecule #2: HmAb64 Fv
| Supramolecule | Name: HmAb64 Fv / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: CNE40 SOSIP trimer
| Supramolecule | Name: CNE40 SOSIP trimer / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: CNE40 SOSIP Envelope glycoprotein gp120
| Macromolecule | Name: CNE40 SOSIP Envelope glycoprotein gp120 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 55.018422 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GNLWVTVYYG VPVWKEATTT LFCASDAKAY DTEVHNVWAT HACVPADPNP QEMLLKNVTE NFNMWKNEMV NQMHEDVISL WDQSLKPCV KLTPLCVTLN CTNVKINSTS NETCIDSGNN STCNETYKEM RNCSFNATTV VRDKQQKMYA LFYKLDIVPL N SGYKNSSD ...String: GNLWVTVYYG VPVWKEATTT LFCASDAKAY DTEVHNVWAT HACVPADPNP QEMLLKNVTE NFNMWKNEMV NQMHEDVISL WDQSLKPCV KLTPLCVTLN CTNVKINSTS NETCIDSGNN STCNETYKEM RNCSFNATTV VRDKQQKMYA LFYKLDIVPL N SGYKNSSD ETYRLINCNT SAITQACPKV SFDPIPIHYC TPAGYALLKC NNKTFNGTGP CNNVSTVQCT HGIKPVVSTQ LL LNGSLAE EEIIIRSENL TNNAKTIIVH LKEPVNITCE RPNNNTRKSI RIGPGQTFYA TGDIIGNIRE AHCNISRSQW NKT LQGVGE KLAELFPNKT IVFKNSSGGD LEITTHSFNC RGEFFYCNTT DLFNSTYWSN GTYITQSNSS SINITLPCRI KQII NMWQE VGRAIYAPPI AGQITCISNI TGLLLLRDGG KEANGTEIFR PGGGDMRDNW RSELYKYKVV EIKPLGVAPT KCRRR VVER RRRRR UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: CNE40 SOSIP Transmembrane protein gp41
| Macromolecule | Name: CNE40 SOSIP Transmembrane protein gp41 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.205479 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASITLTVQA RQLLSGIVQQ QSNLLKAPEA QQHLLQLTVW GIKQLQTRVL AIERYLKDQQ LLGIWGCSG KLICCTAVPW NSSWSNKSQT EIWNNMTWMQ WDREINNYTD IIYRLLEESQ NQQENNEEDL LALD UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #3: HmAb64 Fv heavy chain
| Macromolecule | Name: HmAb64 Fv heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.745402 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE VKKPGASVKV SCKASGYTFT SYDITWVRQA PGQGLEWMGW ISAYNGDTNY AQRLQGRVTM TTDTSTSTAY MELRSLRSD DTAVYYCARA KHTVLVTAMR WFDPWGQGTL VTVSS |
-Macromolecule #4: HmAb64 Fv light chain
| Macromolecule | Name: HmAb64 Fv light chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.852093 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSASVGDRVT ITCRASQSIS NYLNWYQQKP GKAPKLLISA TSSLQSGVPS RFSGSGSGTD FTLTISSLQP DDFATYYCQ QSYSTPWTFG QGTKLEIKRT |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 27 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.0 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 56.76 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 3.74 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 3.3.2) / Number images used: 316221 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)