[English] 日本語
 Yorodumi
Yorodumi- EMDB-26157: Cryo-EM structure of BG505 SOSIP HIV-1 Env trimer in complex with... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV / Env / Broadly Neutralizing / Darpin / CD4 / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationhelper T cell enhancement of adaptive immune response / interleukin-16 binding / interleukin-16 receptor activity / response to methamphetamine hydrochloride / maintenance of protein location in cell / cellular response to ionomycin / T cell selection / MHC class II protein binding / positive regulation of kinase activity / cellular response to granulocyte macrophage colony-stimulating factor stimulus ...helper T cell enhancement of adaptive immune response / interleukin-16 binding / interleukin-16 receptor activity / response to methamphetamine hydrochloride / maintenance of protein location in cell / cellular response to ionomycin / T cell selection / MHC class II protein binding / positive regulation of kinase activity / cellular response to granulocyte macrophage colony-stimulating factor stimulus / interleukin-15-mediated signaling pathway / positive regulation of monocyte differentiation / Nef Mediated CD4 Down-regulation / Alpha-defensins / regulation of T cell activation / response to vitamin D / extracellular matrix structural constituent / Other interleukin signaling / T cell receptor complex / enzyme-linked receptor protein signaling pathway / Translocation of ZAP-70 to Immunological synapse / Phosphorylation of CD3 and TCR zeta chains / positive regulation of protein kinase activity / regulation of calcium ion transport / positive regulation of calcium ion transport into cytosol / macrophage differentiation / Generation of second messenger molecules / immunoglobulin binding / T cell differentiation / Co-inhibition by PD-1 / symbiont-mediated perturbation of host defense response / Binding and entry of HIV virion / coreceptor activity / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of T cell proliferation / positive regulation of calcium-mediated signaling / positive regulation of interleukin-2 production / cell surface receptor protein tyrosine kinase signaling pathway / protein tyrosine kinase binding / host cell endosome membrane / Vpu mediated degradation of CD4 / clathrin-coated endocytic vesicle membrane / calcium-mediated signaling / positive regulation of protein phosphorylation / MHC class II protein complex binding / transmembrane signaling receptor activity / response to estradiol / Downstream TCR signaling / Cargo recognition for clathrin-mediated endocytosis / signaling receptor activity / Clathrin-mediated endocytosis / virus receptor activity / response to ethanol / defense response to Gram-negative bacterium / clathrin-dependent endocytosis of virus by host cell / adaptive immune response / early endosome / positive regulation of viral entry into host cell / cell surface receptor signaling pathway / positive regulation of ERK1 and ERK2 cascade / positive regulation of canonical NF-kappaB signal transduction / cell adhesion / positive regulation of MAPK cascade / immune response / viral protein processing / membrane raft / endoplasmic reticulum lumen / fusion of virus membrane with host plasma membrane / external side of plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / lipid binding / protein kinase binding / endoplasmic reticulum membrane / positive regulation of DNA-templated transcription / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / enzyme binding / signal transduction / protein homodimerization activity / zinc ion binding / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.87 Å | |||||||||
 Authors Authors | Cerutti G / Gorman J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Trapping the HIV-1 V3 loop in a helical conformation enables broad neutralization. Authors: Matthias Glögl / Nikolas Friedrich / Gabriele Cerutti / Thomas Lemmin / Young D Kwon / Jason Gorman / Liridona Maliqi / Peer R E Mittl / Maria C Hesselman / Daniel Schmidt / Jacqueline ...Authors: Matthias Glögl / Nikolas Friedrich / Gabriele Cerutti / Thomas Lemmin / Young D Kwon / Jason Gorman / Liridona Maliqi / Peer R E Mittl / Maria C Hesselman / Daniel Schmidt / Jacqueline Weber / Caio Foulkes / Adam S Dingens / Tatsiana Bylund / Adam S Olia / Raffaello Verardi / Thomas Reinberg / Nicolas S Baumann / Peter Rusert / Birgit Dreier / Lawrence Shapiro / Peter D Kwong / Andreas Plückthun / Alexandra Trkola /   Abstract: The third variable (V3) loop on the human immunodeficiency virus 1 (HIV-1) envelope glycoprotein trimer is indispensable for virus cell entry. Conformational masking of V3 within the trimer allows ...The third variable (V3) loop on the human immunodeficiency virus 1 (HIV-1) envelope glycoprotein trimer is indispensable for virus cell entry. Conformational masking of V3 within the trimer allows efficient neutralization via V3 only by rare, broadly neutralizing glycan-dependent antibodies targeting the closed prefusion trimer but not by abundant antibodies that access the V3 crown on open trimers after CD4 attachment. Here, we report on a distinct category of V3-specific inhibitors based on designed ankyrin repeat protein (DARPin) technology that reinstitute the CD4-bound state as a key neutralization target with up to >90% breadth. Broadly neutralizing DARPins (bnDs) bound V3 solely on open envelope and recognized a four-turn amphipathic α-helix in the carboxy-terminal half of V3 (amino acids 314-324), which we termed 'αV3C'. The bnD contact surface on αV3C was as conserved as the CD4 binding site. Molecular dynamics and escape mutation analyses underscored the functional relevance of αV3C, highlighting the potential of αV3C-based inhibitors and, more generally, of postattachment inhibition of HIV-1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26157.map.gz emd_26157.map.gz | 117.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26157-v30.xml emd-26157-v30.xml emd-26157.xml emd-26157.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26157.png emd_26157.png | 123.2 KB | ||
| Filedesc metadata |  emd-26157.cif.gz emd-26157.cif.gz | 6.3 KB | ||
| Others |  emd_26157_additional_1.map.gz emd_26157_additional_1.map.gz | 118 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26157 http://ftp.pdbj.org/pub/emdb/structures/EMD-26157 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26157 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26157 | HTTPS FTP |
-Validation report
| Summary document |  emd_26157_validation.pdf.gz emd_26157_validation.pdf.gz | 480.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26157_full_validation.pdf.gz emd_26157_full_validation.pdf.gz | 480.4 KB | Display | |
| Data in XML |  emd_26157_validation.xml.gz emd_26157_validation.xml.gz | 6.6 KB | Display | |
| Data in CIF |  emd_26157_validation.cif.gz emd_26157_validation.cif.gz | 7.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26157 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26157 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26157 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26157 | HTTPS FTP |
-Related structure data
| Related structure data |  7txdMC  7z7cC 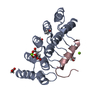 8aedC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26157.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26157.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Local refinement map
| File | emd_26157_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local refinement map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) ...
| Entire | Name: BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9 |
|---|---|
| Components |
|
-Supramolecule #1: BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) ...
| Supramolecule | Name: BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: Envelope glycoprotein gp120
| Macromolecule | Name: Envelope glycoprotein gp120 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 54.064277 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT ...String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT SAITQACPKV SFEPIPIHYC APAGFAILKC KDKKFNGTGP CPSVSTVQCT HGIKPVVSTQ LLLNGSLAEE EV MIRSENI TNNAKNILVQ FNTPVQINCT RPNNNTRKSI RIGPGQAFYA TGDIIGDIRQ AHCNVSKATW NETLGKVVKQ LRK HFGNNT IIRFANSSGG DLEVTTHSFN CGGEFFYCNT SGLFNSTWIS NTSVQGSNST GSNDSITLPC RIKQIINMWQ RIGQ AMYAP PIQGVIRCVS NITGLILTRD GGSTNSTTET FRPGGGDMRD NWRSELYKYK VVKIEPLGVA PTRCKRRVVG RRRRR R UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: Envelope glycoprotein gp41
| Macromolecule | Name: Envelope glycoprotein gp41 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.146482 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEA QQHLLKLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSG KLICCTNVPW NSSWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALD UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #3: T-cell surface glycoprotein CD4
| Macromolecule | Name: T-cell surface glycoprotein CD4 / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 20.50326 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: KKVVLGKKGD TVELTCTASQ KKSIQFHWKN SNQIKILGNQ GSFLTKGPSK LNDRADSRRS LWDQGNFPLI IKNLKIEDSD TYICEVEDQ KEEVQLLVFG LTANSDTHLL QGQSLTLTLE SPPGSSPSVQ CRSPRGKNIQ GGKTLSVSQL ELQDSGTWTC T VLQNQKKV EFKIDIVVLA FQKASNT UniProtKB: T-cell surface glycoprotein CD4 |
-Macromolecule #4: Broadly neutralizing darpin bnd.9
| Macromolecule | Name: Broadly neutralizing darpin bnd.9 / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 21.214891 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GLNDIFEAQK IEWHEGSGGS DLGKKLLEAA YYGQLDEVRI LMANGADVNA VDVHGITPLH LAAYFGHLEI VEVLLKTGAD VNARDNRGI TPLHLAAAMG HLEIVEVLLK AGADVNAFDE AGDTPLHLAA LKGHLEIVEV LLKHGADVNA QDKFGKTAFD I SIDNGNED ...String: GLNDIFEAQK IEWHEGSGGS DLGKKLLEAA YYGQLDEVRI LMANGADVNA VDVHGITPLH LAAYFGHLEI VEVLLKTGAD VNARDNRGI TPLHLAAAMG HLEIVEVLLK AGADVNAFDE AGDTPLHLAA LKGHLEIVEV LLKHGADVNA QDKFGKTAFD I SIDNGNED IAEVLQKAAK LNLEVLFQGP HHHHHHHH |
-Macromolecule #9: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 9 / Number of copies: 30 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.87 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 115325 |
| Initial angle assignment | Type: OTHER / Software - Name: cryoSPARC |
| Final angle assignment | Type: OTHER / Software - Name: cryoSPARC |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)