+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer | |||||||||
 Map data Map data | sharpened | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV-1 / SOSIP / Vaccine / IMMUNE SYSTEM / fusion peptide / FP / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.99 Å | |||||||||
 Authors Authors | Gorman J / Kwong PD | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2023 Journal: J Virol / Year: 2023Title: Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization. Authors: Mallika Sastry / Anita Changela / Jason Gorman / Kai Xu / Gwo-Yu Chuang / Chen-Hsiang Shen / Cheng Cheng / Hui Geng / Sijy O'Dell / Li Ou / Reda Rawi / Mateo Reveiz / Guillaume B E Stewart- ...Authors: Mallika Sastry / Anita Changela / Jason Gorman / Kai Xu / Gwo-Yu Chuang / Chen-Hsiang Shen / Cheng Cheng / Hui Geng / Sijy O'Dell / Li Ou / Reda Rawi / Mateo Reveiz / Guillaume B E Stewart-Jones / Shuishu Wang / Baoshan Zhang / Tongqing Zhou / Andrea Biju / Michael Chambers / Xuejun Chen / Angela R Corrigan / Bob C Lin / Mark K Louder / Krisha McKee / Alexandra F Nazzari / Adam S Olia / Danealle K Parchment / Edward K Sarfo / Tyler Stephens / Jonathan Stuckey / Yaroslav Tsybovsky / Raffaello Verardi / Yiran Wang / Cheng-Yan Zheng / Yuling Chen / Nicole A Doria-Rose / Adrian B McDermott / John R Mascola / Peter D Kwong /  Abstract: While neutralizing antibodies that target the HIV-1 fusion peptide have been elicited in mice by vaccination, antibodies reported thus far have been from only a single antibody class that could ...While neutralizing antibodies that target the HIV-1 fusion peptide have been elicited in mice by vaccination, antibodies reported thus far have been from only a single antibody class that could neutralize ~30% of HIV-1 strains. To explore the ability of the murine immune system to generate cross-clade neutralizing antibodies and to investigate how higher breadth and potency might be achieved, we tested 17 prime-boost regimens that utilized diverse fusion peptide-carrier conjugates and HIV-1 envelope trimers with different fusion peptides. We observed priming in mice with fusion peptide-carrier conjugates of variable peptide length to elicit higher neutralizing responses, a result we confirmed in guinea pigs. From vaccinated mice, we isolated 21 antibodies, belonging to 4 distinct classes of fusion peptide-directed antibodies capable of cross-clade neutralization. Top antibodies from each class collectively neutralized over 50% of a 208-strain panel. Structural analyses - both X-ray and cryo-EM - revealed each antibody class to recognize a distinct conformation of fusion peptide and to have a binding pocket capable of accommodating diverse fusion peptides. Murine vaccinations can thus elicit diverse neutralizing antibodies, and altering peptide length during prime can improve the elicitation of cross-clade responses targeting the fusion peptide site of HIV-1 vulnerability. The HIV-1 fusion peptide has been identified as a site for elicitation of broadly neutralizing antibodies, with prior studies demonstrating that priming with fusion peptide-based immunogens and boosting with soluble envelope (Env) trimers can elicit cross-clade HIV-1-neutralizing responses. To improve the neutralizing breadth and potency of fusion peptide-directed responses, we evaluated vaccine regimens that incorporated diverse fusion peptide-conjugates and Env trimers with variation in fusion peptide length and sequence. We found that variation in peptide length during prime elicits enhanced neutralizing responses in mice and guinea pigs. We identified vaccine-elicited murine monoclonal antibodies from distinct classes capable of cross-clade neutralization and of diverse fusion peptide recognition. Our findings lend insight into improved immunogens and regimens for HIV-1 vaccine development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29836.map.gz emd_29836.map.gz | 117.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29836-v30.xml emd-29836-v30.xml emd-29836.xml emd-29836.xml | 23.7 KB 23.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29836.png emd_29836.png | 108.5 KB | ||
| Masks |  emd_29836_msk_1.map emd_29836_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-29836.cif.gz emd-29836.cif.gz | 7.1 KB | ||
| Others |  emd_29836_additional_1.map.gz emd_29836_additional_1.map.gz emd_29836_half_map_1.map.gz emd_29836_half_map_1.map.gz emd_29836_half_map_2.map.gz emd_29836_half_map_2.map.gz | 27.6 MB 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29836 http://ftp.pdbj.org/pub/emdb/structures/EMD-29836 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29836 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29836 | HTTPS FTP |
-Validation report
| Summary document |  emd_29836_validation.pdf.gz emd_29836_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29836_full_validation.pdf.gz emd_29836_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_29836_validation.xml.gz emd_29836_validation.xml.gz | 14.1 KB | Display | |
| Data in CIF |  emd_29836_validation.cif.gz emd_29836_validation.cif.gz | 16.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29836 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29836 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29836 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29836 | HTTPS FTP |
-Related structure data
| Related structure data |  8g85MC  8fr6C  8g9wC  8g9xC  8g9yC  8gasC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29836.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29836.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0733 Å | ||||||||||||||||||||||||||||||||||||
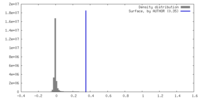
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29836_msk_1.map emd_29836_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
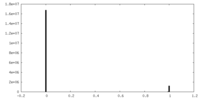
| Density Histograms |
-Additional map: unsharpened
| File | emd_29836_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
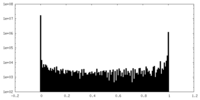
| Density Histograms |
-Half map: half map a
| File | emd_29836_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map a | ||||||||||||
| Projections & Slices |
| ||||||||||||
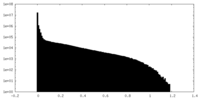
| Density Histograms |
-Half map: half map b
| File | emd_29836_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map b | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer
| Entire | Name: vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer |
|---|---|
| Components |
|
-Supramolecule #1: vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer
| Supramolecule | Name: vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: Envelope glycoprotein gp120
| Macromolecule | Name: Envelope glycoprotein gp120 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 54.086324 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT ...String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT SACTQACPKV SFEPIPIHYC APAGFAILKC KDKKFNGTGP CPSVSTVQCT HGIKPVVSTQ LLLNGSLAEE EV MIRSENI TNNAKNILVQ FNTPVQINCT RPNNNTRKSI RIGPGQAFYA TGDIIGDIRQ AHCNVSKATW NETLGKVVKQ LRK HFGNNT IIRFANSSGG DLEVTTHSFN CGGEFFYCNT SGLFNSTWIS NTSVQGSNST GSNDSITLPC RIKQIINMWQ RIGQ CMYAP PIQGVIRCVS NITGLILTRD GGSTNSTTET FRPGGGDMRD NWRSELYKYK VVKIEPLGVA PTRCKRRVVG RRRRR R UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: Envelope glycoprotein gp41
| Macromolecule | Name: Envelope glycoprotein gp41 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.146482 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEA QQHLLKLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSG KLICCTNVPW NSSWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALD UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #3: vFP52.02 Heavy
| Macromolecule | Name: vFP52.02 Heavy / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.125581 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DVQLQESGPG LVKPSQSLSL TCSVTGYSIT SAYYWNWIRQ FPGKKLEWMG YLLYDGSTGY NPSLKNRISI TRDTSKNQFF LKLNSVTPE DTATYYCSRE GNNRSYWGQG TTLIVSS |
-Macromolecule #4: vFP52.02 Light
| Macromolecule | Name: vFP52.02 Light / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.951338 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVMTQSHRF MSTSVGDRVS ITCKASQSVD TAVAWYQQKP GQSPKLLIYW ASTRHPGVPD RFTGSGSGTD FILTISNVQS EDLADYFCH QFDRYPLTFG DGTKLELK |
-Macromolecule #11: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 11 / Number of copies: 35 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: PBS |
| Grid | Model: C-flat-1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average exposure time: 10.0 sec. / Average electron dose: 69.87 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)