+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | L5A7 Fab bound to Indonesia2005 Hemagglutinin | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | influenza / hemagglutinin / antibody / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral budding from plasma membrane / clathrin-dependent endocytosis of virus by host cell / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Influenza A virus Influenza A virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.59 Å | |||||||||
 Authors Authors | Olia AS / Gorman J / Kwong PD | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Front Immunol / Year: 2024 Journal: Front Immunol / Year: 2024Title: Anti-idiotype isolation of a broad and potent influenza A virus-neutralizing human antibody. Authors: Adam S Olia / Madhu Prabhakaran / Darcy R Harris / Crystal Sao-Fong Cheung / Rebecca A Gillespie / Jason Gorman / Abigayle Hoover / Nicholas C Morano / Amine Ourahmane / Abhinaya Srikanth / ...Authors: Adam S Olia / Madhu Prabhakaran / Darcy R Harris / Crystal Sao-Fong Cheung / Rebecca A Gillespie / Jason Gorman / Abigayle Hoover / Nicholas C Morano / Amine Ourahmane / Abhinaya Srikanth / Shuishu Wang / Weiwei Wu / Tongqing Zhou / Sarah F Andrews / Masaru Kanekiyo / Lawrence Shapiro / Adrian B McDermott / Peter D Kwong /  Abstract: The VH6-1 class of antibodies includes some of the broadest and most potent antibodies that neutralize influenza A virus. Here, we elicit and isolate anti-idiotype antibodies against germline ...The VH6-1 class of antibodies includes some of the broadest and most potent antibodies that neutralize influenza A virus. Here, we elicit and isolate anti-idiotype antibodies against germline versions of VH6-1 antibodies, use these to sort human leukocytes, and isolate a new VH6-1-class member, antibody L5A7, which potently neutralized diverse group 1 and group 2 influenza A strains. While its heavy chain derived from the canonical IGHV6-1 heavy chain gene used by the class, L5A7 utilized a light chain gene, IGKV1-9, which had not been previously observed in other VH6-1-class antibodies. The cryo-EM structure of L5A7 in complex with Indonesia 2005 hemagglutinin revealed a nearly identical binding mode to other VH6-1-class members. The structure of L5A7 bound to the isolating anti-idiotype antibody, 28H6E11, revealed a shared surface for binding anti-idiotype and hemagglutinin that included two critical L5A7 regions: an FG motif in the third heavy chain-complementary determining region (CDR H3) and the CDR L1 loop. Surprisingly, the chemistries of L5A7 interactions with hemagglutinin and with anti-idiotype were substantially different. Overall, we demonstrate anti-idiotype-based isolation of a broad and potent influenza A virus-neutralizing antibody, revealing that anti-idiotypic selection of antibodies can involve features other than chemical mimicry of the target antigen. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43529.map.gz emd_43529.map.gz | 230.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43529-v30.xml emd-43529-v30.xml emd-43529.xml emd-43529.xml | 21 KB 21 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43529.png emd_43529.png | 44.8 KB | ||
| Filedesc metadata |  emd-43529.cif.gz emd-43529.cif.gz | 7 KB | ||
| Others |  emd_43529_half_map_1.map.gz emd_43529_half_map_1.map.gz emd_43529_half_map_2.map.gz emd_43529_half_map_2.map.gz | 226.4 MB 226.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43529 http://ftp.pdbj.org/pub/emdb/structures/EMD-43529 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43529 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43529 | HTTPS FTP |
-Related structure data
| Related structure data |  8vueMC  8vuzC  8vvbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43529.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43529.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.083 Å | ||||||||||||||||||||||||||||||||||||
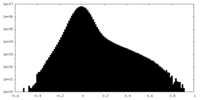
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_43529_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
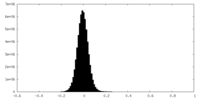
| Density Histograms |
-Half map: #2
| File | emd_43529_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
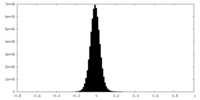
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of L5A7 Fab and Indonesia2005 HA
| Entire | Name: Complex of L5A7 Fab and Indonesia2005 HA |
|---|---|
| Components |
|
-Supramolecule #1: Complex of L5A7 Fab and Indonesia2005 HA
| Supramolecule | Name: Complex of L5A7 Fab and Indonesia2005 HA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Hemagglutinin HA1 chain
| Macromolecule | Name: Hemagglutinin HA1 chain / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Influenza A virus Influenza A virus |
| Molecular weight | Theoretical: 36.792523 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QDQICIGYHA NNSTEQVDTI MEKNVTVTHA QDILEKTHNG KLCDLDGVKP LILRDCSVAG WLLGNPMCDE FINVPEWSYI VEKANPTND LCYPGSFNDY EELKHLLSRI NHFEKIQIIP KSSWSDHEAS SGVSSACPYL GSPSFFRNVV WLIKKNSTYP T IKKSYNNT ...String: QDQICIGYHA NNSTEQVDTI MEKNVTVTHA QDILEKTHNG KLCDLDGVKP LILRDCSVAG WLLGNPMCDE FINVPEWSYI VEKANPTND LCYPGSFNDY EELKHLLSRI NHFEKIQIIP KSSWSDHEAS SGVSSACPYL GSPSFFRNVV WLIKKNSTYP T IKKSYNNT NQEDLLVLWG IHHPNDAAEQ TRLYQNPTTY ISIGTSTLNQ RLVPKIATRS KVNGQSGRME FFWTILKPND AI NFESNGN FIAPEYAYKI VKKGDSAIMK SELEYGNCNT KCQTPMGAIN SSMPFHNIHP LTIGECPKYV KSNRLVLATG LRN SPQRES UniProtKB: Hemagglutinin |
-Macromolecule #2: Hemagglutinin HA2 chain
| Macromolecule | Name: Hemagglutinin HA2 chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Influenza A virus Influenza A virus |
| Molecular weight | Theoretical: 19.640781 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: RRKKRGLFGA IAGFIEGGWQ GMVDGWYGYH HSNEQGSGYA ADKESTQKAI DGVTNKVNSI IDKMNTQFEA VGREFNNLER RIENLNKKM EDGFLDVWTY NAELLVLMEN ERTLDFHDSN VKNLYDKVRL QLRDNAKELG NGCFEFYHKC DNECMESIRN G TYNYPQYS E UniProtKB: Hemagglutinin |
-Macromolecule #3: L5A7 Fab Heavy Chain
| Macromolecule | Name: L5A7 Fab Heavy Chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.642494 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLQQSGPG LVKPSQTLSL TCGISGDSVS SDAAAWDWIR QSPSRGLEWL GRTFYRSRWH HDYSESVKNR ITINADTSKN QFSLQLTSV TPEDTATYYC ARAGVRVFGI IVNSLDYWGQ GTLVTVSSAS TKGPSVFPLA PSSKSTSGGT AALGCLVKDY F PEPVTVSW ...String: QVQLQQSGPG LVKPSQTLSL TCGISGDSVS SDAAAWDWIR QSPSRGLEWL GRTFYRSRWH HDYSESVKNR ITINADTSKN QFSLQLTSV TPEDTATYYC ARAGVRVFGI IVNSLDYWGQ GTLVTVSSAS TKGPSVFPLA PSSKSTSGGT AALGCLVKDY F PEPVTVSW NSGALTSGVH TFPAVLQSSG LYSLSSVVTV PSSSLGTQTY ICNVNHKPSN TKVDKKVEPK SC |
-Macromolecule #4: L5A7 Fab Light Chain
| Macromolecule | Name: L5A7 Fab Light Chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.716301 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AIQLTQSPSS LSASVGDRVT ITCRASQATS SYLAWYQQKP GKAPKLLIYA ASTLQSGVPS RFSGSGSGTD FTLTITSLQP EDFATYYCQ LSKTFGPGTK VEIKRTVAAP SVFIFPPSDE QLKSGTASVV CLLNNFYPRE AKVQWKVDNA LQSGNSQESV T EQDSKDST ...String: AIQLTQSPSS LSASVGDRVT ITCRASQATS SYLAWYQQKP GKAPKLLIYA ASTLQSGVPS RFSGSGSGTD FTLTITSLQP EDFATYYCQ LSKTFGPGTK VEIKRTVAAP SVFIFPPSDE QLKSGTASVV CLLNNFYPRE AKVQWKVDNA LQSGNSQESV T EQDSKDST YSLSSTLTLS KADYEKHKVY ACEVTHQGLS SPVTKSFNRG EC |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: C-flat / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 64.12 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)